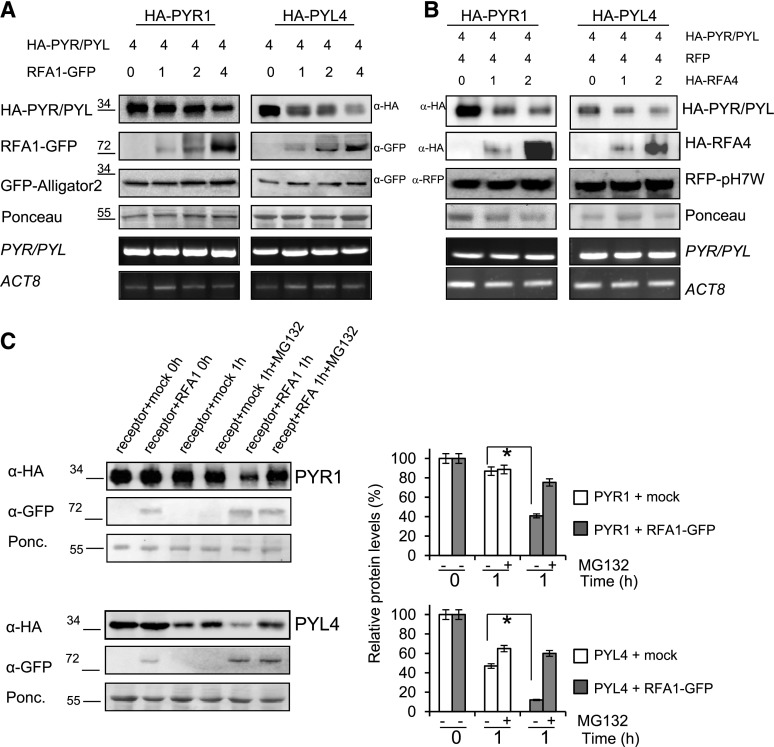
Figure 5.
Analysis of RFA1- and RFA4-promoted degradation of PYR1 and PYL4 by in vivo and semi-in vivo assays. A and B, In vivo degradation of either PYR1 or PYL4 was observed in coinfiltration experiments with increasing amounts of RFA1 or RFA4. The ratio of the relative concentration of agrobacteria used in the different coinfiltrations is indicated by numbers (top). In A, cell extracts were analyzed using anti-HA to detect HA-tagged PYR1/PYL4 and anti-GFP to detect GFP-RFA1, whereas in B, anti-HA was used to detect both HA-tagged PYR1/PYL4 and HA-RFA4. Anti-GFP or anti-RFP antibodies were used to detect the internal control of GFP or RFP, respectively. The mRNA expression levels of the target PYR1 and PYL4 genes and ACTIN8 (ACT8) were analyzed by reverse transcription-semiquantitative PCR. Molecular masses of marker proteins are indicated in kilodaltons. C, Cell-free experiments show HA-tagged PYR1/PYL4 degradation promoted by RFA1. Degradation of either HA-PYL4 or HA-PYR1 was performed by mixing N. benthamiana cell extracts from separated agroinfiltrations. HA-PYL4 or HA-PYR1 extract was mixed with RFA1-GFP extract or mock control extract and incubated at 4°C during 1 h either in the absence or presence of MG132. Proteins were detected by immunoblot as described above. Ponceau staining of the Rubisco protein is shown as a loading control. Graphs at right show the quantification of the experiment. Values are averages ± sd obtained from three independent experiments (n = 3 leaf extracts in each experiment). *, P < 0.05 (Student’s t test) compared with the corresponding mock-treated sample.