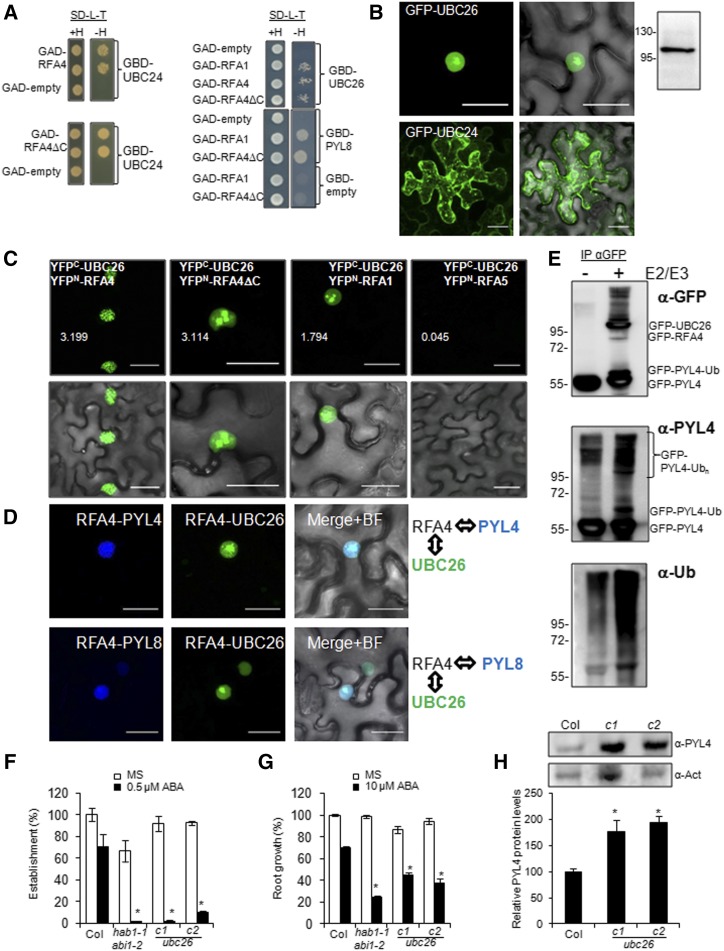
Figure 6.
UBC26 and the UBC26-RFA4-ABA receptor complex are localized in the nucleus. A, Y2H interactions of RFA1 and RFA4. Transformed yeast cultures were grown overnight in liquid synthetic defined (SD) medium lacking Leu and Trp, and a dilution (10−1) of these cultures was dropped on either control medium lacking Leu and Trp (SD–Leu–Trp+His) or selective medium additionally lacking His (SD–Leu–Trp–His). Empty vectors were used as negative controls, and yeast was allowed to grow for 3 d at 30°C before interaction was scored. B, GFP-UBC26 localizes in the nucleus upon transient expression in N. benthamiana, whereas GFP-UBC24 seems to be membrane associated. Immunoblotting analysis using anti-GFP was used to verify the expression of GFP-UBC26. C, BiFC interaction of RFA4 and UBC26 in the nucleus of N. benthamiana epidermal cells. Confocal images show transiently transformed leaf cells coexpressing YFPC-UBC26 and YFPN-RFA4, RFA4ΔC, RFA1, or RFA5. Note that the E2-E3 interaction decorates nuclear speckles, whereas the PYL4-E3 interaction is absent in the nucleolus (Fig. 2A). Deletion of the acidic domain of RFA4 changes the nuclear pattern of the E2-E3 interaction compared with RFA4FL. YFPC-UBC26 does not interact with YFPN-RFA5, which is another member of the RFA family. Quantification of YFP signal was analyzed by ImageJ software, and the arbitrary units of fluorescence are indicated in numbers. D, Multicolor BiFC reveals the formation of E2-E3-PYL4/8 nuclear complexes. Confocal images show transiently transformed N. benthamiana epidermal cells coexpressing SCFPC-RFA4, SCFPN-PYL4/8, and VENUSN-UBC26. The RFA4-PYL4/8 interaction was visualized through reconstitution of the SCFP, whereas the RFA4-UBC26 interaction gave rise to SCFPC-VENUSN fluorescent protein. SCFPC-RFA4 coexpressed with SCFPN or VENUSN did not reconstitute fluorescent protein (Supplemental Fig. S3B). Constructs were delivered into N. benthamiana epidermal leaves through A. tumefaciens-mediated transfection. Leaves were examined using confocal laser scanning microscopy 48 to 72 h after infiltration. Bars = 30 μm in B to D. E, In vivo ubiquitination of PYL4 by UBC26-RFA4 in N. benthamiana leaf cells. Agrobacteria encoding GFP-PYL4 were coinfiltrated in N. benthamiana cells with agrobacteria lacking or encoding GFP-UBC26/GFP-RFA4 proteins (−/+ E2E3). Protein extracts were immunoprecipitated using anti-GFP and next immunoblotted with anti-GFP, anti-PYL4, and anti-Ub to detect ubiquitination of GFP-PYL4 in the absence or presence of GFP-UBC26/GFP-RFA4 proteins. Extraction of GFP-RFA4 for SDS-PAGE analysis seems to be inefficient because of the large C-terminal acidic domain. F, Enhanced sensitivity to ABA-mediated inhibition of seedling establishment. Approximately 100 seeds of each genotype (three independent experiments) were sown on Murashige and Skoog (MS) plates lacking or supplemented with 0.5 µm ABA and scored for the presence of both green cotyledons and the first pair of true leaves after 7d. Values are averages ± sd. *, P < 0.01 (Student’s t test) with respect to wild-type Columbia-0 (Col-0) assayed in the same conditions. G, Enhanced sensitivity to ABA-mediated inhibition of root growth. Quantification is shown for ABA-mediated inhibition of root growth in the indicated genotypes compared with the wild type. *, P < 0.01 (Student’s t test) with respect to wild-type Col-0 assayed in the same conditions. H, Accumulation of endogenous PYL4 protein in ubc26-c1 and ubc26-c2 alleles compared with the wild type. Seedlings of the indicated genotypes were grown for 7 d in MS medium, and total proteins were extracted and subjected to immunoblot analysis using the indicated antibodies. Actin was analyzed as a protein-loading control, and relative PYL4 protein levels were quantified. Values are averages ± sd obtained from three independent experiments (n > 20 seedlings in each experiment). *, P < 0.05 (Student’s t test) compared with wild-type Col-0.