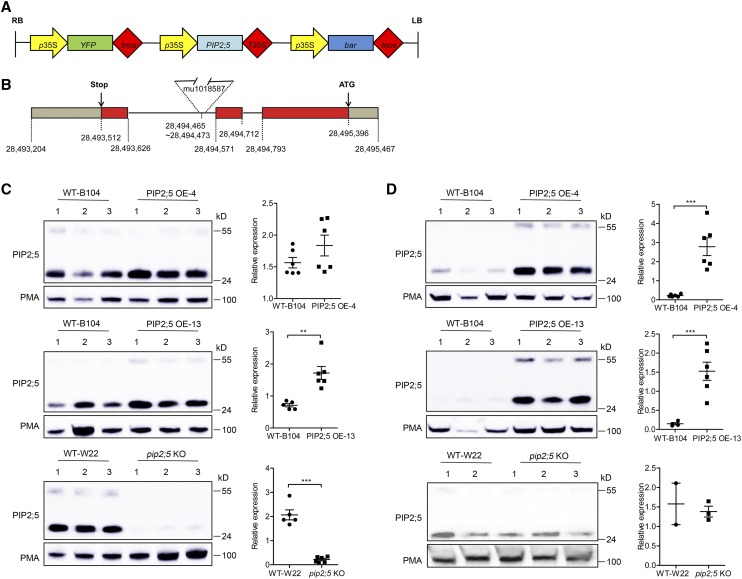
Figure 1.
PIP2;5 protein levels in the maize lines deregulated in its expression. A, Schematic representation of the transfer DNA used to overexpress PIP2;5 in the B104 maize line. YFP, Yellow fluorescent proteins; tnos, terminator of the nopaline gene; bar, gene conferring resistance to bialaphos. B, Genomic structure of the PIP2;5 gene with the position of the Mu transposon insertion in the pip2;5 KO line (W22 background). The data source of gene position and insertion site is from “Maize B73 RefGen_v3” in MaizeGDB (https://maizegdb.org/). The PIP2;5 exons are in red. C and D, PIP2;5 protein level in root (C) and leaf (D) in the wild type (WT; indicated by wild-type B104 and wild-type W22), two PIP2;5 OE lines (OE-4 and OE-13), and a pip2;5 knockout line (pip2;5 KO). The plants were cultured under hydroponic conditions and the microsomal fractions were extracted from primary roots and leaves of 1-week-old seedlings. Proteins (20 μg [C] or 30 μg [D]) were subjected to western blotting using antibodies raised against PIP2;5 or PMA (H+−ATPases). The PMA signal was used to control the gel loading and normalize the PIP2;5 signals (plots at right). In the quantification panels, data are expressed as the mean ± se of two independent experiments, each experiment containing two to three plants for each maize line. Significant differences among the treatments are indicated by **P < 0.01 and ***P < 0.001.