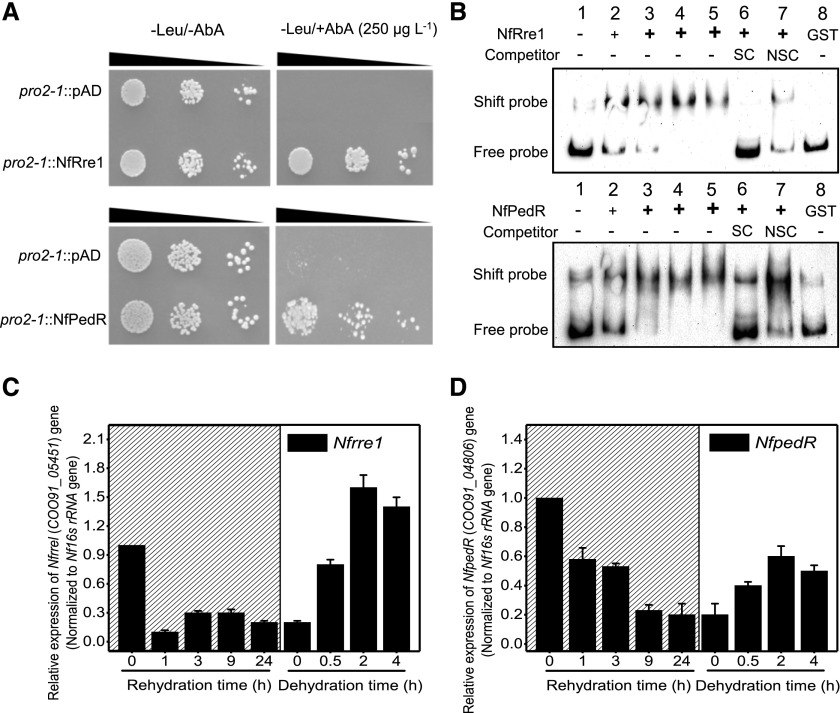
Figure 7.
Identification of transcriptional regulators binding to the promoter of NfdnaK2. A, Verification of interactions between the cis-acting element Promoter2-1 (pro2-1) and putative transcription factors NfRre1 or NfPedR by Y1H assay. The transformed yeast cells (10 μL) with DNA fragments and regulator proteins at OD600 0.1, 0.01, and 0.001 (indicated by the slope of the black triangles from left to right) were tested on SD/-Leu medium with or without 250 μg L−1 AbA. B, Interaction analyses between Promoter2-1 and NfRre1 or NfPedR by EMSA. Biotin-labeled Promoter2-1 (5 ng) was incubated with increasing concentrations of purified GST-tagged NfRre1 or NfPedR (lane 1, 0 ng; lane 2, 100 ng; lane 3, 200 ng; lane 4, 400 ng; and lane 5, 600 ng). The unlabeled Promoter2-1 (lane 6, 500 ng) and poly(dI-dC) (lane 7, 2,000 ng) were used as specific competitor (SC) and nonspecific competitor (NSC) inhibitors, respectively. GST-tag (lane 8, 5 ng Biotin-labeled Promoter2-1 was incubated with 200 ng GST protein, which was the equal amount as GST-tagged NfRre1 or NfPedR used in lanes 6 and 7) was used as a negative control to exclude the nonspecific binding of NfRre1/NfPedR with promoter2-1. The shift probe indicates the biotin-labeled promoter2-1 interacting with NfRre1 or NfPedR, and the free probe is the biotin-labeled promoter2-1 without interacting with NfRre1 or NfPedR. C and D, Relative transcriptional levels of Nfrre1 (C) or NfpedR (D) during rehydration and subsequent dehydration. The dried field-collected N. flagelliforme samples were rehydrated (marked by hatching) for 0, 1, 3, 9, and 24 h and subsequently dehydrated for 0.5, 2, and 4 h. Each treatment was repeated three times independently, and data are shown as the mean ± sd of three independent replicates.