Figure 1. Overview of molecular networks and single-cell network modeling approaches.
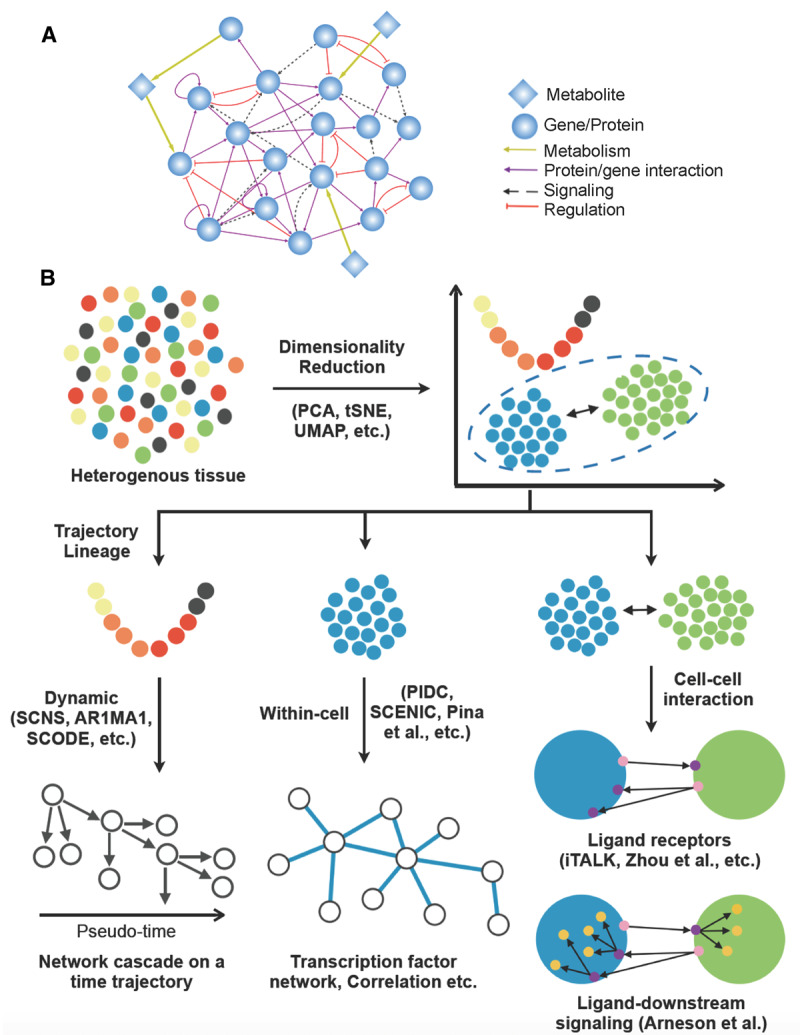
(A) Various types of network nodes and edges (connections) in a molecular network. (B) Concepts and example methods of current single-cell network modeling approaches. Transcriptomes of single cells from tissue samples are first processed and clustered using dimension reduction techniques, followed by cell identity determination using known cell markers. Cell populations in various dynamic states can be ordered using pseudo-time and trajectory analyses, and the pseudo-time information can be used in dynamic network modeling. Gene networks within a given cell population and cell–cell communication networks can also be reconstructed based on various assumptions and algorithms.
