Figure 3.
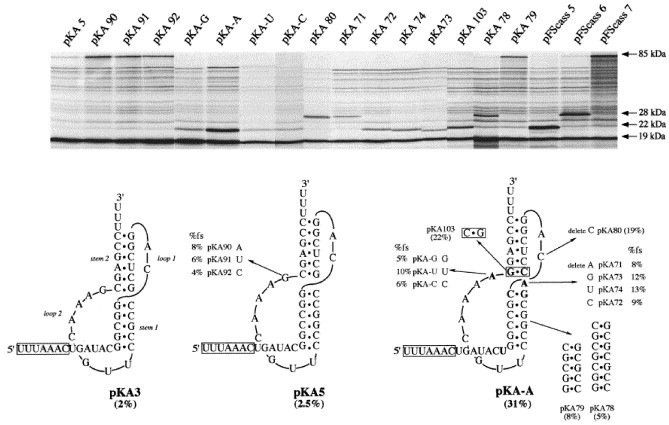
Analysis of the pKA-A frameshift signal by mutagenesis. This Figure shows the primary sequence and proposed secondary structure of the pKA3, pKA5 and pKA-A pseudoknots. The slippery sequence is boxed. Nucleotides of pKA-A that differ in orientation or presence from those of pKA5 are shown in bold. A series of mutations were created in pKA5 and pKA-A, and the effects on frameshifting measured by in vitro transcription and translation in the RRL. The upper portion of the Figure shows the RRL translation products synthesised in response to mRNAs derived from BamHI-digested plasmids. Products were labelled with [35S]methionine, separated on a SDS-15 % polyacrylamide gel and detected by autoradiography. The frameshifted (22, 28 or 85 kDa) and non-frameshifted (19 kDa) species are marked with arrows. The size of the −1 frameshift product produced by the various mutant RNAs is determined by the number of nucleotides in the pseudoknot (see Results). The predicted sizes are 22 kDa (pKA-G, A, 69, 70, 72, 73, 74, 103 pFScass 5), 28 kDa (pKA71, 78, 80, pFScass 6) or 85 kDa (pKA5, 79, 90, 91, 92, pFScass 7). pFScass 5, 6 and 7 contain the minimal IBV frameshift signal (Brierley et al., 1992) and were translated to mark the position of the 22, 28 and 85 kDa frameshift products. In translations of pKA-based constructs in RRL, a low level of background polypeptides are present which arise as a result of aberrant initiation events (see Brierley et al., 1992). These can potentially introduce an inaccuracy of up to 2 % in estimations of frameshift efficiency and we define a non-functional construct as one displaying a frameshift efficiency of ⩽2 %.
