Abstract
Growing resistant wheat (Triticum aestivum L) varieties is an important strategy for the control of leaf rust, caused by Puccinia triticina Eriks. This study sought to identify the chromosomal location and effects of leaf rust resistance loci in five Canadian spring wheat cultivars. The parents and doubled haploid lines of crosses Carberry/AC Cadillac, Carberry/Vesper, Vesper/Lillian, Vesper/Stettler and Stettler/Red Fife were assessed for leaf rust severity and infection response in field nurseries in Canada near Swift Current, SK from 2013 to 2015, Morden, MB from 2015 to 2017 and Brandon, MB in 2016, and in New Zealand near Lincoln in 2014. The populations were genotyped with the 90K Infinium iSelect assay and quantitative trait loci (QTL) analysis was performed. A high density consensus map generated based on 14 doubled haploid populations and integrating SNP and SSR markers was used to compare QTL identified in different populations. AC Cadillac contributed QTL on chromosomes 2A, 3B and 7B (2 loci), Carberry on 1A, 2B (2 loci), 2D, 4B (2 loci), 5A, 6A, 7A and 7D, Lillian on 4A and 7D, Stettler on 2D and 6B, Vesper on 1B, 1D, 2A, 6B and 7B (2 loci), and Red Fife on 7A and 7B. Lillian contributed to a novel locus QLr.spa-4A, and similarly Carberry at QLr.spa-5A. The discovery of novel leaf rust resistance QTL QLr.spa-4A and QLr.spa-5A, and several others in contemporary Canada Western Red Spring wheat varieties is a tremendous addition to our present knowledge of resistance gene deployment in breeding. Carberry demonstrated substantial stacking of genes which could be supplemented with the genes identified in other cultivars with the expectation of increasing efficacy of resistance to leaf rust and longevity with little risk of linkage drag.
Introduction
Leaf rust, caused by Puccinia triticina Eriks., is an economically devastating fungal pathogen threatening wheat (Triticum aestivum L.) production worldwide [1–4]. Leaf rust occurs more regularly and in more regions world-wide than stem rust (P. graminis) or stripe rust (P. striiformis) of wheat [5]. Many studies indicate that P. triticina spores travel long distances by wind or man, and cause damage to wheat beyond their country of origin. For example, in North America several studies [2, 5, 6] indicated the disease establishes in the fall on winter wheats that are grown in the southern USA, and travels by winds the following spring and summer to the northern USA and Canada along the “Puccinia pathway”.
Growing resistant wheat varieties is an important method for control of leaf rust by farmers because input costs are minimized with reduced requirement of fungicides while environmental sustainability is improved [3]. However, achieving durable resistance can be difficult as the rust pathogen continues to evolve and overcome major genes that have been deployed [2, 7]. In addition to evolution of new races, exotic incursions of rust pathogen races have occurred in recent decades and pose a threat to wheat production areas on different continents [2].
Two types of resistance, seedling (or all-stage) resistance and adult plant resistance (APR), to wheat rusts are known [8]. Most seedling resistance genes are effective from the early seedling stage throughout the life of the plant and are characterized by race specificity and low infection types; whereas adult plant resistance is largely effective at the adult plant growth stage [3, 8]. Seedling resistance is typically monogenic and has been favoured in breeding because of the high level of expressivity and simplicity of phenotypic selection, but most genes have been overcome by the emergence of virulent races. This is illustrated in the case of spring wheat in Canada. The most common leaf rust resistance genes in Canada Western Red Spring (CWRS) wheat varieties are Lr1, Lr10, Lr13, Lr14a, Lr16, Lr21 and Lr34 [3, 9]. Renown was the first wheat variety to be widely grown in Canada having the resistance gene Lr14a, but virulence to the gene was common by 1945 due to changes in the P. triticina population [3]. Subsequently Lr16 was deployed but lost its effectiveness [3]. Next the Lr21 gene, which has been cloned [10], succumbed to virulence in 2010 in the United States and in Canada in 2011 [11]. Furthermore genes Lr1, Lr10 and Lr13 are no longer effective [12]. Ultimately the majority of known all-stage resistance genes have been defeated [13–15] leaving few genes for deployment in resistance breeding.
Adult plant resistance genes such as Lr34, Lr46 and Lr67 [13–15] have also been used along with resistance from seedling genes. Other well characterized leaf rust adult plant resistance genes include Lr68, Lr74, Lr75, Lr77, and Lr78 [16]. In some cases, these genes work synergistically with seedling resistance such as Lr10, Lr13, Lr16, and Lr18 which confer resistance in combination with other genes, particularly Lr34 [3, 17]. Combinations of adult plant resistance genes and other minor effect genes that condition resistance to a broad spectrum of P. triticina races are key to the development of wheat varieties with long-lasting resistance to leaf rust [18].
Mapping resistance genes in existing adapted parental stocks is necessary in order to understand the effective gene combinations and for efficient marker assisted selection [19]. Recent molecular mapping studies uncovered several quantitatively inherited sources of leaf rust resistance based on minor effect genes in different wheat germplasm. For example, in their review of research from the last fifteen years, Li et al. [20] documented 80 leaf rust resistance QTL involving sixteen wheat chromosomes. A few years later, in their review work, Da Silva, Zanella [21] reported 249 leaf rust resistance QTL identified in 70 bi-parental populations and 79 donor lines. Modern Canadian spring wheat cultivars Lillian, Stettler, Carberry, AC Cadillac, Vesper and a founder cultivar Red Fife show varying levels of leaf rust resistance, and although Singh et al. [22] studied leaf rust resistance in cultivars Carberry and AC Cadillac, the genes providing resistance have not been fully characterized. This study sought to identify the chromosomal locations and effects of genes controlling leaf rust resistance in spring wheat cultivars Lillian, Stettler, Carberry, AC Cadillac, Vesper and Red Fife.
Materials and methods
Plant materials
Five doubled haploid (DH) populations—Carberry/AC Cadillac, Carberry/Vesper, Vesper/Lillian, Vesper/Stettler and Stettler/Red Fife—were generated by the maize pollen method [23] from F1 plants of crosses between cultivars of the market class Canada Western Red Spring (CWRS), including a founder cultivar Red Fife. The pedigree descriptions and leaf rust resistance genes possibly possessed by these parents are described in S1 Table. The number of lines evaluated in each of the five populations ranged from 94 to 775 (S2 Table). The number of lines phenotyped per population varied for different reasons mainly based on the number of lines available from the doubled haploid system, cost of genotyping, or taking advantage of phenotyping a subset of the population used for a different study. The lines used for genotyping were chosen randomly and became the basis of the population size for QTL analysis.
Rust infection phenotyping under field conditions
Testing of the populations was done as described by Bokore et al. [24] and Singh et al. [22]. Briefly, the populations were grown in un-replicated single row plots and assessed for leaf rust severity and infection response in disease nurseries in Canada near Swift Current, SK from 2013 to 2015, Morden, MB from 2015 to 2017, Brandon, MB in 2016 and in New Zealand near Lincoln in 2014 (S2 Table). Parents and check cultivars were repeated in each experiment and spreader rows of susceptible genotypes were planted around the plots to enhance disease development.
Inoculum of P. triticina was generated by increasing urediniospores of all races in the proportions that they were found in western Canada in the year prior to the field trial. The frequency of virulence to 16 leaf rust resistance genes in these population mixes is shown in Table 1. Urediniospores of these multi-race mixtures were used to inoculate spreader rows susceptible to leaf rust at the Swift Current, Morden and Brandon locations. This inoculum was generated by increasing and collecting urediniospores from a representative mixture of the virulence phenotypes found in Canada during the annual national virulence survey in the previous year [25]. For each year, all the isolates generated during the virulence survey of Manitoba and Saskatchewan were combined to generate this field inoculum. Each year, 18 to 64 unique virulence phenotypes were included in this field inoculum (Table 1). In a given season, the same P. triticina race composition was used in Morden, Swift Current and Brandon trials. At the Morden and Brandon locations urediniospores were suspended in light mineral oil (Soltrol, Chevron Phillips Chemical Co) and sprayed on the leaves of the spreader rows at early tillering, subsequently leaf rust developed on the spreader rows and urediniospores were windblown to the test lines to provide infection. At Swift Current, spreader rows of susceptible genotypes were needle inoculated with urediniospores. At Lincoln, natural infection was the sole source of inoculum. No artificial inoculation was carried out.
Table 1. Frequency of virulence to 16 leaf rust resistance genes in the Puccinia triticina inoculum mixture used to inoculate field screening nurseries in Canada between 2010 and 2016.
| Year | Number of isolates | Number of virulence phenotypes | Gene | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lr1 | Lr2a | Lr2c | Lr3 | Lr9 | Lr16 | Lr24 | Lr26 | Lr3ka | Lr11 | Lr17 | Lr30 | LrB | Lr10 | Lr14a | Lr18 | Lr21 | |||
| 2010 | 341 | 18 | 100 | 48.1 | 49.9 | 100 | 31.7 | 0 | 60.4 | 10 | 18.2 | 1.5 | 51.9 | 10.6 | 51.9 | 100 | 78.3 | 0 | 0 |
| 2011 | 216 | 33 | 100 | 52.3 | 63.6 | 100 | 12.1 | 2.8 | 60.7 | 13.1 | 20.1 | 0.9 | 47.7 | 15.4 | 46.7 | 99.5 | 57.9 | 0 | 7 |
| 2012 | 177 | 28 | 100.0 | 62.1 | 67.8 | 100.0 | 24.9 | 4.0 | 52.0 | 7.3 | 14.1 | 0.0 | 38.4 | 10.2 | 40.1 | 100.0 | 47.5 | 2.3 | 10.2 |
| 2013 | 236 | 29 | 100 | 42.8 | 43.2 | 100 | 33.5 | 0 | 41.1 | 10.6 | 28 | 5.9 | 56.8 | 26.3 | 56.8 | 96.2 | 66.9 | 0 | 10.6 |
| 2014 | 93 | 29 | 100 | 55.8 | 62.1 | 98.9 | 28.4 | 2.1 | 24.2 | 13.7 | 24.2 | 7.4 | 62.1 | 24.2 | 46.3 | 93.7 | 45.3 | 1.1 | 2.1 |
| 2015 | 208 | 42 | 99 | 41.3 | 41.8 | 99.5 | 53.8 | 4.3 | 48.6 | 24.5 | 37 | 5.3 | 62.5 | 36.5 | 63.9 | 98.1 | 73.1 | 0.5 | 10.1 |
| 2016 | 233 | 64 | 100 | 24.9 | 26.2 | 100 | 51.5 | 16.7 | 63.1 | 29.2 | 47.6 | 3.9 | 73.8 | 45.9 | 77.7 | 98.7 | 83.7 | 0 | 4.3 |
The percent leaf rust severity of infected flag leaves was scored using the modified Cobb Scale [26] at all locations except Lincoln in 2014 where a scale of 0 to 10 was used and converted to percent by multiplying values by 10. Infection response was recorded as resistant (R), resistant to moderately resistant (RMR), moderately resistant (MR), mesothetic (X), moderately resistant to moderately susceptible (MRMS), moderately susceptible (MS), moderately susceptible to susceptible (MSS), and susceptible (S). Infection response was not recorded for trials planted at Lincoln. The infection response scores were converted into numeric values based on R = 1, RMR = 2, MR = 3, X = 4, MRMS = 5, MS = 6, MSS = 7, and S = 8 for QTL analysis.
Genotyping, construction of linkage maps and QTL analysis
The DNA of parents and DH lines was extracted from young leaves with the BioSprint 96 DNA Plant Kit (QIAGEN Science, Maryland, USA). Table 2 shows the number of DH lines genotyped with the 90K Infinium iSelect SNP wheat assay (Illumina Inc., San Diego, CA). In addition, SNP12, a co-dominant Lr34 diagnostic SNP marker modified from the dominant marker caSNP12 [27] on chromosome 7D was integrated into the maps of Carberry/Vesper and Vesper/Lillian populations. SNP12 is defined with primer sequences AAG AAT GAA GCC TCC GAA TG (forward) and CAT TCA GTC ACC TCG CAG (reverse). The SNP12 assay was performed on the Roche LightCycler II Real-Time Thermal Cycler using the High Resolution Melt (HRM) module (S3 Table). During PCR, a 117 base amplicon is amplified containing the SNP nucleotide. Because the amplicon is monomorphic in size for all samples, polymorphism cannot be detected via electrophoresis. Therefore, the additional step of HRM analysis is performed to detect the differences in melt curve signatures caused by the differing SNP base after PCR is complete.
Table 2. Number of lines, number of linkage groups, number of markers, genomic size and map density of five doubled haploid mapping populations used in the leaf rust resistance QTL analysis.
| Population name | Number of doubled haploid lines | Number of linkage groups | Number of markers | Length (cM) | Density |
|---|---|---|---|---|---|
| Carberry/AC Cadillac | 775 | 29 | 6806 | 3237.9 | 0.7 |
| Carberry/Vesper | 188 | 28 | 6138 | 1835.4 | 0.3 |
| Vesper/Lillian | 283 | 29 | 7839 | 3679.5 | 0.8 |
| Vesper/Stettler | 94 | 22 | 4989 | 2002.0 | 0.4 |
| Stettler/Red Fife | 218 | 26 | 9983 | 3247.6 | 0.3 |
Genetic maps were built for each of the five populations, using the two-step mapping strategy described previously [28, 29]. Briefly, ‘draft’ linkage maps for individual populations were generated using the minimum spanning tree map (MSTMap) software using a stringent cut off p-value of 1E-10 and a maximum distance between markers of 15 cM. Then, the ‘draft’ maps were refined using the MapDisto version 1.7.5 software using a cut off recombination value of 35%, a minimum LOD score of 3.0 and the Kosambi mapping function. The best order of markers was generated using both “AutoCheckInversions” and “AutoRipple” commands. Then, we built a consensus map based on fourteen hexaploid wheat mapping populations including the five QTL populations (Carberry/AC Cadillac, Carberry/Vesper, Vesper/Lillian, Stettler/Vesper and Stettler/Red fife) used in the current leaf rust resistance mapping study and nine others, namely: 8021V2/AC Karma [30], AAC Concord/CDC Hughes, Attila/CDC Go [31], Carberry/Thatcher, Cutler/AC Barrie [28], Norstar/Capelle Despres [29], Norstar/Manitou [29], Norstar/Winter Manitou [29] and RL4452/AC Domain. In addition to SNP markers, 8021V2/AC Karma was genotyped with 529 microsatellite or simple sequence repeat (SSR) [32–34] markers. The individual genetic maps were integrated into a consensus map, using the LPmerge R package [35, 36]. This software uses a linear programming algorithm to minimize the mean absolute error between the consensus map and the individual maps. For the goodness-of-fit for the consensus map, LPmerge computes a root-mean-square error (RMSE) per linkage group by comparing the position (in cM) of all markers on the consensus map with that on the individual maps. This metric was calculated for different maximum interval sizes (k in the algorithm), ranging from 1 to 10. The value of k minimizing the mean RMSE per linkage group was selected for construction of the consensus map. The consensus map integrated both SNP and SSR markers.
Quantitative trait loci analysis (QTL) was applied to all population x environment combinations for leaf rust severity and infection response except for Carberry/Vesper at Lincoln in 2014 in which disease development was insufficient to discriminate among the lines. QTL analyses were performed on five rust evaluated bi-parental crosses whereas the consensus map based on fourteen populations was used only for comparing QTL positions. The leaf rust resistance loci were identified by performing QTL analysis using MapQTL.6® [37]. The permutation test option (1000 permutations) within MapQTL was applied to determine the significant threshold for the logarithm of the odds (LOD). Genome-wide threshold levels were used to declare significant QTL at the 5% level of significance. Automatic co-factor detection based on backward elimination to identify the co-factor markers as well as manual co-factor selection was performed for Multiple QTL Mapping (MQM). The position in the genetic map of the SNP markers associated with each of the QTL were aligned with the hexaploid consensus map using MapChart [38] to investigate (1) the relationship of the QTL identified in different cultivars, and (2) if the QTL are located in the same region with those reported by other studies.
A microsatellite marker wmc44 associated with the leaf rust gene Lr46 [39, 40] and co-located to chromosome 1BL, was run on Carberry/Vesper and Vesper/Lillian populations to see the similarity of a QTL identified on chromosome 1BL and derived from Vesper in the Vesper/Lillian population. As Lr46 was originally reported in Pavon 76 and Lalbahadur (a monosomic line carrying Lr46 [15]), the two lines were genotyped by a Kompetitive Allele Specific PCR (KASP) marker converted from SNP marker probes associated with the 1BL QTL in Vesper in the Vesper/Lillian population to investigate if the 1BL QTL corresponds with the Lr46 gene.
Results
Leaf rust reaction
The response to leaf rust of parents of the populations varied in different environments (Fig 1, S4 Table). The greatest amount of leaf rust occurred at Morden in 2015. In this environment, Lillian was the most resistant cultivar among the parents and displayed 3% disease severity, while Red Fife with a severity of 78% was the most susceptible. The highest reaction scored on Carberry at 15% severity, Vesper at 30% and Stettler at 60% was observed in the Morden 2015 environment. The Carberry/AC Cadillac population was not planted at Morden, but Carberry displayed lower disease severity than AC Cadillac in the Swift Current environments with adequate levels of disease. The infection responses of the cultivars paralleled the severities, with Lillian, Vesper, Carberry and AC Cadillac varying between R and MRMS, and that of Stettler and Red Fife varying from MR to S.
Fig 1. Frequency distribution of five doubled populations (a) Vesper/Lillian, (b) Vesper/Stettler, (c) Carberry/Vesper, (d) Stettler/Red Fife, and (e) Carberry/AC Cadillac for adult plant leaf rust severity.
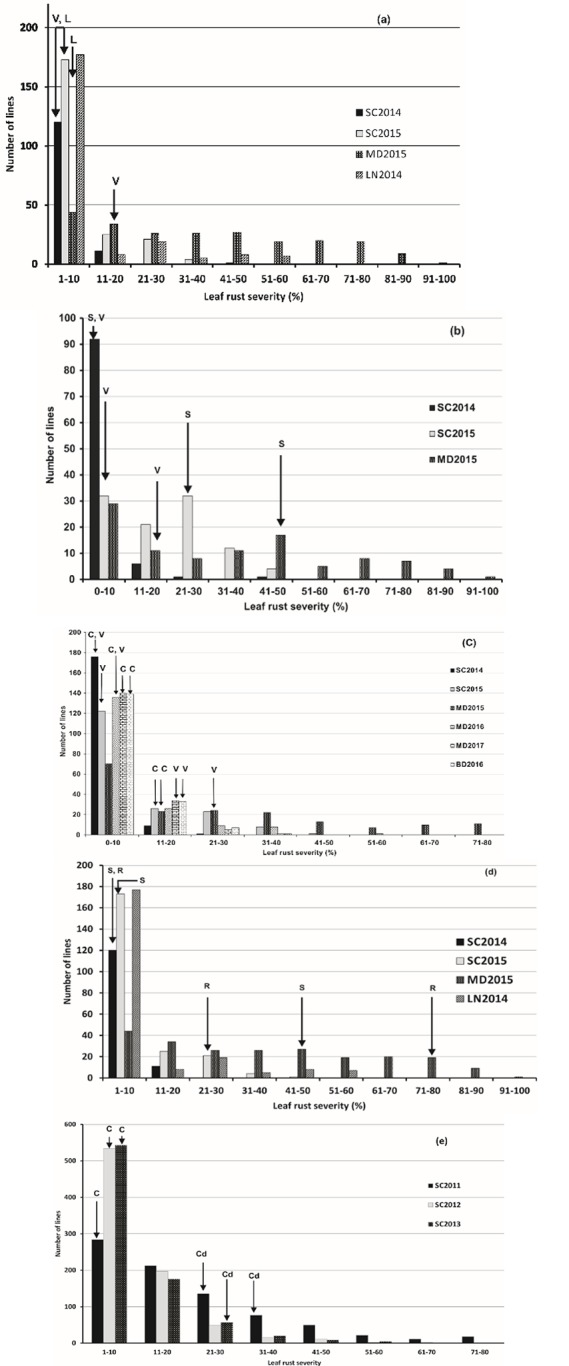
Arrows indicate parent leaf rust severity: C, Carberry; Cd, AC Cadillac; V, Vesper; L, Lillian; S, Stettler; Rf, Red Fife. In the key, the test year is preceded by the location defined as follows: SC, Swift Current, MD, Morden, Canada, and LN, Lincoln, New Zealand.
The distributions of leaf rust severity were continuous for all the populations, and lines transgressive for resistance appeared occasionally in the resistant tail and regularly in the susceptible tail (Fig 1). The populations generally displayed skewed distributions with a preponderance of lines showing resistance to leaf rust. Considering all the test environments, the greatest range in severity of the lines was with the Vesper/Lillian (1–95%), Vesper/Stettler (1–95%) and Stettler/Red Fife (1–95%) populations, while the Carberry/Vesper was somewhat narrower (0–80%) as was Carberry/AC Cadillac (1–80%).
Genetic maps
Individual high-density maps were built for the five populations phenotyped for leaf rust in this QTL mapping study. The number of markers, genomic size as map length in cM, and map density of the populations used for consensus map development are shown in S5 Table and for leaf rust QTL analysis in Table 2 along with number of linkage groups generated for the latter. For example, for the Vesper/Lillian population, the map consisted of 29 linkage groups and 7839 markers that spanned 3679.5 cM. The consensus map we built comprising of SNP and SSR markers using 14 mapping populations allowed comparison of our research results with previous reports on leaf rust resistance genes.
S1 File presents the hexaploid wheat consensus map and genetic maps of the 14 different populations used to build the consensus map. Table 3 demonstrates some of the features of the hexaploid wheat consensus map. It consisted of 36715 SNP and SSR markers spanning all hexaploid wheat chromosomes and covered a length of 3162 cM with an average marker spacing of 0.13 cM/marker. The D genome covered the smallest map length at 642 cM, with the lowest average marker density of 6.4 markers/cM, while A genome marker density was 11.3 markers/cM and the B genome 14.6 markers/cM.
Table 3. Statistics characterizing totals as well as individual chromosomes of the consensus map built from 14 hexaploid wheat populationsa: Chromosome name, number of markers, map length (cM), map density and average marker spacing.
| Chromosome | Number of markers | Length (cM) | Map density (cM/Marker) | Marker density (Marker/cM) |
|---|---|---|---|---|
| 1A | 2526 | 186.37 | 0.07 | 13.6 |
| 2A | 1978 | 187.69 | 0.09 | 10.5 |
| 3A | 1673 | 194.13 | 0.12 | 8.6 |
| 4A | 1630 | 169.63 | 0.1 | 9.6 |
| 5A | 1795 | 218.33 | 0.12 | 8.2 |
| 6A | 2685 | 151.0 | 0.06 | 17.8 |
| 7A | 2502 | 198.55 | 0.08 | 12.6 |
| Total A Genome | 14789 | 1305.7 | 0.09 | 11.3 |
| 1B | 2450 | 166.95 | 0.07 | 14.7 |
| 2B | 3849 | 199.06 | 0.05 | 19.3 |
| 3B | 2371 | 154.36 | 0.07 | 15.4 |
| 4B | 1294 | 153.77 | 0.12 | 8.4 |
| 5B | 3018 | 197.45 | 0.07 | 15.3 |
| 6B | 2702 | 206.8 | 0.08 | 13.1 |
| 7B | 2108 | 137.29 | 0.07 | 15.4 |
| Total B Genome | 17792 | 1215.68 | 0.08 | 14.6 |
| 1D | 762 | 85.85 | 0.11 | 8.9 |
| 2D | 1291 | 119.22 | 0.09 | 10.8 |
| 3D | 886 | 122.46 | 0.14 | 7.2 |
| 4D | 120 | 71.98 | 0.6 | 1.7 |
| 5D | 412 | 69.11 | 0.17 | 6.0 |
| 6D | 398 | 83.39 | 0.21 | 4.8 |
| 7D | 265 | 89.51 | 0.34 | 3.0 |
| Total D Genome | 4134 | 641.52 | 0.24 | 6.4 |
| Total | 36715 | 3162.9 | 0.13 | 11.6 |
a The 14 populations are derived from crosses between 8021V2/AC Karma, AAC Concord/CDC Hughes, Attila/CDC Go, Carberry/Thatcher, Cutler/AC Barrie, Norstar/Capelle Despres, Norstar/Manitou, Norstar/Winter Manitou and RL4452/AC Domain
QTL analysis
Twenty leaf rust resistance QTL were identified in the five populations. In S1 Fig, the positions of the QTL in each population were aligned with the hexaploid wheat consensus map we generated. A summary of markers with the highest LOD scores at each QTL, phenotypic variation explained, and associated additive effects is presented in Table 4. The source of resistance and environments that revealed the QTL are listed in S6 Table, whereas the detailed information on each QTL is given in S7 Table.
Table 4. Closest marker, associated LOD score, percentage of phenotypic variation explained by individual QTL and the level of additive effect of QTL identified in five doubled haploid populations evaluated for their field responses against leaf rust in nurseries near Swift Current, SK, Morden and Brandon, MB, Canada and Lincoln, New Zealand.
| QTL | Closest marker | Position, cM | Traita | LODb | PVE(%)c | Addd | LOD | PVE(%) | Add | LOD | PVE(%) | Add | LOD | PVE(%) | Add | LOD | PVE(%) | Add | LOD | PVE(%) | Add | Sourcee |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Carberry/AC Cadillac (4f) | Swift Current 2011 | Swift Current 2012 | Swift Current 2013 | Swift Current 2014 | ||||||||||||||||||
| QLr.spa-1A | IACX1465 | 42.7 | Sev | 8.4 | 4.8 | 9.6 | 15.1 | 8.6 | 7.3 | 11.6 | 6.7 | 7.1 | C | |||||||||
| IR | 3.4 | 2.0 | 0.6 | 3.8 | 2.2 | 0.5 | 4.0 | 2.3 | 0.4 | C | ||||||||||||
| QLr.spa-2A.1 | BS00041816_51 | 50.0 | Sev | 4.6 | 2.7 | -2.9 | 4.2 | 2.4 | -1.6 | 7.1 | 4.1 | -2.3 | Cd | |||||||||
| IR | 10.2 | 1.8 | -0.3 | 10.1 | 5.8 | -0.2 | 7.2 | 4.2 | -0.3 | Cd | ||||||||||||
| QLr.spa-2B.1 | Excalibur_c39493_251 | 7.4 | Sev | 3.4 | 2.0 | 4.7 | C | |||||||||||||||
| IR | 3.3 | 1.9 | 0.3 | C | ||||||||||||||||||
| QLr.spa-2B.2 | Kukri_c53810_137 | 38.8 | Sev | 6.4 | 3.6 | 8.3 | 10.6 | 5.9 | 6.1 | 15.5 | 8.7 | 8.2 | C | |||||||||
| IR | 3.5 | 2.0 | 0.6 | 3.8 | 2.2 | 0.2 | 3.3 | 1.8 | 0.4 | C | ||||||||||||
| QLr.spa-2D.1 | Ex_c2115_3369 | 81.7 | Sev | 3.9 | 2.3 | 2.7 | 3.4 | 2.0 | 1.5 | C | ||||||||||||
| IR | ||||||||||||||||||||||
| QLr.spa-3B | Tdurum_contig79629_538 | 15.1 | Sev | 2.1 | 1.2 | -2.0 | 4.0 | 2.3 | -1.7 | Cd | ||||||||||||
| IR | 2.6 | 1.5 | -0.2 | 4.8 | 2.8 | -0.2 | Cd | |||||||||||||||
| QLr.spa-4B.1 | Tdurum_contig12204_1131 | 0.0 | Sev | 3.4 | 1.9 | 2.5 | 3.3 | 1.9 | 1.4 | 3.5 | 2.0 | 1.6 | C | |||||||||
| IR | 3.0 | 1.7 | 0.2 | 5.3 | 3.0 | 0.3 | 4.5 | 2.5 | 0.2 | C | ||||||||||||
| QLr.spa-4B.2 | BS00021984_51 | 75.7 | Sev | 4.5 | 2.6 | 2.9 | 3.3 | 1.9 | 1.4 | 3.0 | 1.7 | 1.5 | C | |||||||||
| IR | 5.2 | 3.0 | 0.3 | 6.3 | 3.6 | 0.3 | 6.7 | 3.8 | 0.3 | C | ||||||||||||
| QLr.spa-5A | BobWhite_c1387_798 | 41.6 | Sev | 4.5 | 2.6 | 3.1 | 7.9 | 4.6 | 2.3 | 9.4 | 5.4 | 2.8 | 3.8 | 7.3 | 1.8 | C | ||||||
| IR | 4.8 | 2.8 | 0.2 | 3.1 | 1.8 | 0.1 | 6.5 | 3.8 | 0.3 | C | ||||||||||||
| QLr.spa-6A | BobWhite_c39821_195 | 4.2 | Sev | 11.5 | 6.6 | 6.4 | 14.7 | 8.3 | 8.0 | C | ||||||||||||
| QLr.spa-7A | BS00063860_51 | 193.3 | Sev | 3.8 | 2.2 | 1.7 | C | |||||||||||||||
| QLr.spa-7B.1 | Ex_c101666_634 | 31.5 | Sev | 9.2 | 5.2 | -4.1 | 15.5 | 8.6 | -3.0 | 18 | 9.9 | -3.6 | 2.8 | 4.5 | -1.4 | Cd | ||||||
| IR | 12.4 | 7.0 | -0.5 | 14.9 | 8.5 | -0.3 | 18.2 | 10.2 | -0.4 | 5.8 | 10.7 | -0.4 | Cd | |||||||||
| QLr.spa-7B.2 | RAC875_c57326_85 | 145.9 | Sev | 4.5 | 2.4 | -7.4 | 21.3 | 10.6 | -8.8 | 15.2 | 7.6 | -8.3 | Cd | |||||||||
| IR | 3.0 | 1.6 | -0.4 | Cd | ||||||||||||||||||
| Carberry/Vesper (7g) | Swift Current 2014 | Swift Current 2015 | Morden 2015 | Morden 2016 | Morden 2017 | Brandon 2016 | ||||||||||||||||
| QLr.spa-1D | RAC875_c2070_566 | 67.5 | Sev | 6 | 13.8 | 1.6 | 6.7 | 16 | 4.5 | 9.7 | 21.9 | 11 | 2.9 | 7.1 | 1.9 | V | ||||||
| IR | 5 | 11.6 | 0.4 | 5.5 | 12 | 1.3 | 8.4 | 19.4 | 1.2 | V | ||||||||||||
| QLr.spa-2A.2 | Kukri_c46040_620 | 0.0 | Sev | 3.6 | 8.8 | 2.1 | V | |||||||||||||||
| IR | 4.7 | 11.3 | 0.9 | V | ||||||||||||||||||
| QLr.spa-2B.1 | BobWhite_c12144_216 | 0.0 | Sev | 2.5 | 6.1 | -5.8 | 2.8 | 6.8 | -2.8 | 3.6 | 8.8 | -2.2 | 2.5 | 6.3 | -1.7 | C | ||||||
| IR | 2.7 | 6.7 | -0.4 | 3.3 | 8.2 | -0.5 | C | |||||||||||||||
| QLr.spa-7A | BS00053365_51 | 99.8 | Sev | 3.0 | 2.4 | -1.1 | 7.6 | 3.0 | -3.1 | C | ||||||||||||
| IR | 2.6 | 2.6 | -0.7 | C | ||||||||||||||||||
| QLr.spa-7D | SNP12 | 2.0 | Sev | 7.5 | 17.5 | -9.8 | 5.9 | 14 | -4 | 9.9 | 22.4 | -3.5 | 8 | 18.5 | -3 | C | ||||||
| IR | 3.3 | 7.9 | -0.3 | 3.6 | 8.7 | -0.8 | 6.5 | 15.4 | -1.1 | 5.4 | 12.9 | -0.6 | 7.7 | 18 | -0.7 | 7 | 16.5 | -0.6 | C | |||
| Vesper/Lillian (5) | Swift Current 2013 | Swift Current 2014 | Swift Current 2015 | Morden 2015 | Lincoln 2014 | |||||||||||||||||
| QLr.spa-1B | wsnp_Ex_c1058_2020681 | 181.9 | Sev | 2.2 | 3.7 | 2.2 | V | |||||||||||||||
| IR | 3 | 4.9 | 0.5 | |||||||||||||||||||
| QLr.spa-1D | Kukri_c2408_784 | 1.0 | Sev | 23.5 | 34.1 | 8.4 | 13.2 | 20 | 5.1 | 4.4 | 6.9 | 3.4 | 22.7 | 31.1 | 17.5 | 21.9 | 30.4 | 1.3 | V | |||
| IR | 16 | 24.7 | 1.2 | 15.8 | 24 | 1.2 | 15.5 | 22.6 | 1.7 | 19.6 | 27.5 | 1.7 | V | |||||||||
| QLr.spa-4A | Ex_c70424_465 | 73.3 | Sev | 3.8 | 4.1 | -3.1 | 3 | 5 | -2.6 | 4.7 | 7 | -4.7 | 3.3 | 5.2 | -20 | 6.1 | 9 | -0.8 | L | |||
| IR | 4.3 | 6.6 | -1.7 | 4.4 | 6.6 | -0.7 | 3.6 | 5.8 | -0.9 | 3.6 | 5.6 | -2.1 | L | |||||||||
| QLr.spa-6B | BobWhite_c36415_378 | 118.6 | Sev | 3.8 | 6.6 | 3.7 | 4.7 | 7.7 | 3.2 | 3.9 | 6.2 | 0.6 | V | |||||||||
| QLr.spa-7B.1 | Kukri_c109962_396 | 0.0 | Sev | 4.2 | 3.7 | 6.7 | V | |||||||||||||||
| IR | 3 | 5.1 | 0.6 | V | ||||||||||||||||||
| QLr.spa-7B.2 | RFL_Contig71_386 | 232 | Sev | 4.9 | 7.7 | 4.1 | 5.1 | 3.3 | 8.1 | 4.8 | 7.3 | 3.5 | 6.5 | 9.8 | 9.8 | V | ||||||
| IR | 5 | 7.6 | 0.9 | V | ||||||||||||||||||
| QLr.spa-7D | SNP12 | 7.5 | Sev | 5.9 | 9.9 | -4.6 | 3.4 | 5.7 | -2.7 | 3.1 | 5 | -0.8 | 8.4 | 12.9 | -11 | L | ||||||
| IR | 10 | 16.3 | -1 | 6.4 | 10 | -1 | L | |||||||||||||||
| Vesper/Stettler (5) | Swift Current 2014 | Swift Current 2015 | Morden 2015 | Lincoln 2014 | ||||||||||||||||||
| QLr.spa-1D | BobWhite_c4303_524 | 58.6 | S | 5.1 | 22.4 | 3.1 | 7.2 | 29.6 | 8.5 | 12.5 | 44.8 | 18.9 | 3.3 | 15.1 | 5.4 | V | ||||||
| IR | 4.2 | 19.2 | 0.9 | 4.6 | 23.5 | 1.1 | 6.7 | 28 | 1.5 | V | ||||||||||||
| QLr.spa-2A.2 | BS00022393_51 | 50.1 | Sev | 3.5 | 15.7 | 11.2 | V | |||||||||||||||
| IR | 3.4 | 13.6 | 0.7 | 3.2 | 14.4 | 1.1 | V | |||||||||||||||
| Stettler/Red Fife (4) | Swift Current 2014 | Swift Current 2015 | Morden 2015 | Lincoln 2014 | ||||||||||||||||||
| QLr.spa-2D.2 | Kukri_rep_c105822_804 | 59.1 | Sev | 3.3 | 10.5 | 1.1 | 12.9 | 24 | 4.8 | 21.1 | 36.1 | 16 | 6.8 | 13.3 | 0.5 | S | ||||||
| IR | 14.3 | 26.2 | 1 | 22.5 | 37.9 | 1.7 | S | |||||||||||||||
| QLr.spa-6B | BS00010993_51 | 111.1 | Sev | 3.9 | 8 | 2.8 | S | |||||||||||||||
| QLr.spa-7A | tplb0031i24_1212 | 4.4 | Sev | 5.6 | 11.2 | -8.9 | R | |||||||||||||||
| IR | 4.1 | 8.4 | -0.8 | R | ||||||||||||||||||
| QLr.spa-7B.2 | BS00108630_51 | 166 | Sev | 3.1 | 6.2 | -0.3 | R | |||||||||||||||
a Sev = leaf rust severity, IR = leaf rust infection response
b Logarithm of odds (LOD) score
c Percentages of phenotypic variation explained by individual QTL
d Additive effect
e Source of resistance allele: Cd, AC Cadillac; C, Carberry; L, Lillian; V, Vesper; S, Stettler; R, Red Fife
f Total number of test environments per population
g Tested in New Zealand in 2014 but no trait differential
Location in the International Wheat Genome Consortium (IWGC) Chinese spring wheat RefSeq. genome v.1.0 of the SNP markers that detected the leaf rust resistance QTL in the present study is presented in S8 Table. The QTL revealed from the results of mapping will be further elaborated upon on a population by population basis.
Carberry/AC Cadillac population
Genetic mapping of the Carberry/AC Cadillac population resulted in the identification of 13 QTL—nine of them contributed by Carberry on chromosomes 1A (QLr.spa-1A), 2B (2 loci) (QLr.spa-2B.1 and QLr.spa-2B.2), 2D (QLr.spa-2D.1), 4B (2 loci) (QLr.spa-4B.1 and QLr.spa-4b.2), 5A (QLr.spa-5A), 6A (QLr.spa-6A) and 7A (QLr.spa-7A) and four by AC Cadillac on 2A (QLr.spa-2A.1), 3B (QLr.spa-3B) and 7B (2 loci) (QLr.spa-7B.1 and QLr.spa-7B.2) (S1 Fig; Table 4 S6 and S7 Tables). The QTL QLr.spa-1A and QLr.spa-2A.1 were detected in three of four environments in Canada. QLr.spa-1A was among the most effective loci explaining phenotypic variation approaching 9% in leaf rust severity although the infection response explained was lower at 2.3%. On the consensus map, several SSR markers (barc28, gwm164, wmc278, wmc469, and gwm357) are located nearby the SNP markers (IACX1465 and Excalibur_c46833_204) that define QLr.spa-1A. QLr.spa-2A.1 had more impact on infection response explaining up to 6% of the phenotypic variation than leaf rust severity at 4%. SNP markers anchoring QLr.spa-2A.1 mapped in the region of barc5, gwm294 and wmc1709.
Two apparent QTL were detected on chromosome 2B from Carberry. The first, QLr.spa-2B.1 mapped adjacent to SSR markers wwmc661, wmc382b, wmc764, wmc489a and barc35 (S1 Fig) and was detected in only two of four environments. This QLr.spa-2B.1 allele from Carberry was additionally detected in the Carberry/Vesper population. The second 2B QTL, QLr.spa-2B.2, was detected in three out of four Canadian environments. QLr.spa-2B.2 could be quite effective, accounting for up to 9% of the total variation in leaf rust severity although only 2% in infection response. The closet SNP marker to QLr.spa-2B.1, Kukri_c53810_137, was located 0.2–0.9 cM from SSR loci wmc257, wmc25a and wmc154.
The 2D QTL, QLr.spa-2D.1, was significantly associated with leaf rust severity in two environments, but not with infection response. Located on the long arm of the chromosome, this QTL is different from a major QTL, QLr.spa-2D.2, that was identified in Stettler on 2DS about 33 cM from QLr.spa-2D.1. Like QLr.spa-2D.1, the 3B QTL QLr.spa-3B was expressed in two environments albeit marginally in the one and was flanked by UMN10 [41], gwm389, bar147 and xsts3B-142.
QLr.spa-4B.1 and QLr.spa-4B.2 were about 75 cM from each other on chromosome 4B. Both QTL expressed in the same three of four Canadian environments. The explained phenotypic variation for these two QTL was relatively low ranging from 1.7% to 3.8%. Even more consistent than the 4B loci, QLr.spa-5A was detected in all four test environments with a moderately high phenotypic variation of up to 7.3% explained for leaf rust severity. In contrast, QLr.spa-6A was significant in two of the four tests and QLr.spa-7A in a single environment only. Although the effect of QLr.spa-7A was quite low, explaining 2.2% of leaf rust severity, QLr.spa-6A was more effective, explaining up to 8% of the severity.
QLr.spa-7B.1 and QLr.spa-7B.2 corresponded with two QTL detected in the Vesper/Lillian population. QLr.spa-7B.2 was similarly detected in Red Fife in the Stettler/Red Fife population. Although both QTL displayed consistent and quite strong effects in the Carberry/AC Cadillac population, the response was more variable in the Vesper and Red Fife genetic backgrounds. The SNP markers that tagged QLr.spa-7B.1 mapped with SSR marker loci wmc323, wmc606, gwm537 and wmc76, while QLr.spa-7B.2 associated SNP markers mapped with wmc581 and gwm344b (S1 Fig).
Carberry/Vesper population
Five leaf rust resistance QTL were detected in the Carberry/Vesper population, from Carberry on chromosomes 2B (QLr.spa-2B.2), 7A (QLr.spa-7A) and 7D (QLr.spa-7D) and Vesper on 1D (QLr.spa-1D) and 2A (QLr.spa-2A.2) (S1 Fig; Table 4 S6 and S7 Tables). QLr.spa-1D was located in the same genomic region as the Vesper QTL detected in the other two Vesper populations Vesper/Lillian and Vesper/Stettler. The QLr.spa-1D associated SNP markers were located close to three SSR markers (wmc432, barc149 and gdm33b) on the consensus map. The QTL was detected in four out of six Canadian environments and had a strong effect explaining up to 22% of the total variation in leaf rust severity and 20% in infection response. The other Vesper QTL, QLr.spa-2A.2, was also detected in the Vesper/Stettler population. QLr.spa-2A.2 expressed in two out of six Canadian environments and was a moderately strong QTL explaining up to 9% of variation in leaf rust severity and 11% in infection response.
QLr.spa-2B.1 was located to the same region in the Carberry/Vesper population as in the Carberry/AC Cadillac population. It was reasonably stable, detected in four of six Canadian environments, and moderately well expressed with explained variation close to 9% in disease severity and 8% in infection response.
QLr.spa-7A, a relatively weakly expressed QTL, was revealed in two out of six environments. The same resistance QTL was detected, also from Carberry, in the Carberry/AC Cadillac population and from Red Fife in the Stettler/Red Fife population. The third consistent and relatively strongly expressed QTL from Carberry, QLr.spa-7D, was associated with the SNP12 marker and was similarly detected in Lillian, but no segregation occurred between Carberry and AC Cadillac at the locus.
Vesper/Lillian population
Seven leaf rust resistance QTL segregated between Lillian and Vesper; two of them were contributed by Lillian on chromosomes 4A (QLr.spa-4A) and 7D (QLr.spa-7D) while five were contributed by Vesper on 1B (QLr.spa-1B), 1D (QLr.spa-1D), 6B (QLr.spa-6B), and 7B (QLr.spa-7B.1 and QLr.spa-7B.2) (S1 Fig; Table 4, S6 and S7 Tables). QLr.spa-1B, although expressed in only a single Canadian environment, explained a reasonable amount the phenotypic variation at 3.7% of the leaf rust severity and 4.9% of the infection response. The peak marker for QLr.spa-1B, Wsnp_Ex_c1058_2020681, was about 19 cM from SSR marker wmc44 and 3 cM from gwm328. The single marker assay and analysis using the Lr46 marker, wmc44, generated a significant marker-trait association. Interestingly, the assay made with the KASP marker associated the QLr.spa-1B in Vesper produced the same allele among Vesper, Pavon 76, and Lalbahadur.
QLr.spa-1D was a consistent QTL being expressed in all five test environments which included Canada and New Zealand. It exhibited a major effect explaining phenotypic variation of up to 45% for leaf rust severity and 28% for infection response. Like QLr.spa-1D, QLr.spa-4A was also effective in all environments. The QTL spanned a large interval devoid of markers. Associated with SSR markers such as wmc491 and wmc680, the SNP markers CAP11_c279_66, tplb0022j01_1046 and Ex_c70424_465 were located on one flank of the QLr.spa-4A QTL (S1 Fig).
QLr.spa-7B.2 expressed moderately strongly with close to 10% of the leaf rust severity explained and consistently in four of five environments while QLr.spa-7B.1 expressed less strongly and in only a single Canadian environment. The SNP markers within QLr.spa-7B.1 were associated with SSR markers wmc323, wmc606, gwm537 and wmc76 whereas QLr.spa-7B.2 with gwm146 and gwm344b (S1 Fig). QLr.spa-7D was observed in the same environments as QLr.spa-7B.2 which did not include the single year of testing in New Zealand. Maximum expression was greater than that of QLr.spa-7B.2. As previously mentioned, QLr.spa-7D was associated with SNP12.
Vesper/Stettler population
Two leaf rust resistance QTL, including a major QTL on chromosome 1D (QLr.spa-1D) and a minor QTL on 2A (QLr.spa-2A.2) were contributed by Vesper (S1 Fig; Table 4, S6 and S7 Tables). However, no QTL was detected from the second parent, Stettler. Similar to the Vesper/Lillian and Carberry/Vesper populations, the QLr.spa-1D from Vesper/Stettler expressed in all the four tests involving Canada and New Zealand. The QTL explained a considerably large amount of phenotypic variation of up to 45% in leaf rust severity and 28% in infection response. Like in the other two Vesper derived populations, QLr.spa-1D associated SNP markers mapped close to three SSR markers wmc432, barc149 and gdm33b on the consensus map. The QTL on chromosome 2A, QLr.spa-2A.2, although inconsistently detected (two of four environments in Canada) explained substantial phenotypic variation approaching 16% for leaf rust severity and 14% for infection response. The QTL was defined by SNP markers that were located at or near SSR markers wmc407, wmc667, gwm296a and wmc636b (S1 Fig).
Stettler/Red Fife population
Four QTL, one with consistent major effects and three inconsistent with moderate effects on the disease traits, segregated in this population. A QTL on chromosome 2D, QLr.spa-2D.2, and one on 6B, QLr.spa-6B, were contributed by Stettler. The other two, one on chromosome 7A, QLr.spa-7A, and one on 7B, QLr.spa-7B.2, were contributed by Red Fife (S1 Fig; Table 4, S6 and S7 Tables). QLr.spa-2D.2 consistently reduced leaf rust in Canada and New Zealand with at times strong expression explaining up to 38% of the infection response. This QTL was not detected in a cross of Stettler with Vesper. The other three QTL expressed in single environments at moderate strength: QLr.spa-6B and QLr.spa-7A in Canada, and QLr.spa-7B.2 in New Zealand. The Stettler 6B QTL had a similar level of expression as the Vesper allele in the Vesper/Lillian population. The two QTL lay in close proximity, with the Ex_c7101_596 a SNP marker associated with the Stettler QTL located 10 cM from the BobWhite_c36415_378 marker that flanks the QTL from Vesper. The markers at the peak of the QTL were close to SSR markers wmc398, barc24 and wmc182a. Likewise, the QLr.spa-7A derived from Red Fife was in a similar region to the 7A QTL from Carberry in the Carberry/Vesper and Carberry/AC Cadillac populations. The Red Fife QTL, QLr.spa-7B.2, was located in a similar region to the 7B QTL contributed by AC Cadillac and Vesper.
Discussion
The results of QTL analysis were consistent with the continuous phenotypic distributions exhibited by the populations. The skewed distributions with a preponderance of leaf rust resistant progenies indicated multiple resistance factors were segregating. Twenty QTL were detected, demonstrating the genetic potential of the five adapted Canadian spring wheat cultivars as sources of leaf rust resistance. In some cases, lack of segregation indicated resistance genes were shared between two parents, for instance QLr.spa-2A.2 segregated in Carberry/Vesper and Vesper/Stettler but not in Vesper/Lillian indicating Vesper and Lillian share the same resistance allele. Conversely the resistant allele is not present in Carberry and Stettler. The potential for improved resistance through targeted gene deployment of these QTL comes with minimal to no risk of linkage drag.
The alignment of the QTL maps with the consensus map (S1 Fig) assisted in defining the QTL detected in different varieties and populations. The inclusion of both SSR markers along with SNP markers in our consensus map facilitated the comparisons made between the QTL identified in the present study and previous mapping reports. Other published consensus maps [42, 43] were generated based on SNP markers only.
The majority of the QTL were limited to single parental cultivars as the source of resistance except those located on chromosomes 6B, 7A, 7D and the two on 7B which had two or more cultivars as sources. The discussion from this point on will focus on the QTL as they relate to a specific parental cultivar with associations between cultivars being considered when appropriate.
QTL detected in a single cultivar
Carberry resistance
Carberry was attributed with the highest number of leaf rust resistance QTL in this study. The QTL detected only in Carberry include QLr.spa-1A, QLr.spa-2B.1, QLr.spa-2B.2, QLr.spa-2D.1, QLr.spa-4B.1, QLr.spa-4B.2, QLr.spa-5A, and QLr.spa-6A. Flanked by IACX1465 and Excalibur_c46833_204 [42], the 1A QTL, QLr.spa-1A is located on chromosome 1AS. The spring wheat variety Superb, an immediate parent of Carberry, is known to possess Lr10 [44] similarly located on 1AS [45]. Lr10 is one of the widely deployed genes in the western Canadian spring wheat varieties [9, 45–47]. QLr.spa-1A maps in the same region as the Lr10 gene, but the relationship between this gene and QTL remains to be determined.
Although our results suggest the two QTL on chromosome 2B from Carberry represent distinct genes, the associated SNP markers place both on the short arm of the chromosome. Flanking markers for QLr.spa-2B.1 are located adjacent to two SSR markers, wmc661 and wmc764, which are markers on 2BS for Lr16 [48, 49]. Consequently Lr16 is a candidate to explain QLr.spa-2B.1. The second QTL, QLr.spa-2B.2, is next to wmc154 which corresponds with a complex locus responsible for leaf, stripe and stem rust resistance and expression of pseudo-black chaff [22, 50]. Apart from Lr16, Carberry could possess genes such as Lr13 and Lr23 similarly located on chromosome 2BS through its immediate parent Alsen [51, 52]. QLr.spa-2B.1 and QLr.spa-2B.2 were expressed in the majority of tests at a moderate level indicating their usefulness for continued deployment in resistance breeding.
In the present study, we identified two consistent QTL on chromosome 4B, QLr.spa-4B.1 and QLr.spa-4B.2, contributed by Carberry. This finding is in partial agreement with Singh et al. [22] who reported only one QTL from Carberry using DArT markers on a subset of the lines from the population we used in the current study. Referring to associated SNP markers QLr.spa-4B.1 could be placed on 4BS, and QLr.spa-4B.2 on 4BL [42]. A leaf rust resistance locus similar to QLr.spa-4B.1 was reported in the Swiss wheat variety Forno [53]. The closest markers for the Carberry QTL were located within 1 cM distance of gwm368, a marker for the Forno gene. Chromosome 4BL of wheat harbours genes such as Lr12 and Lr31 [54] and Lr49 [55]; but further work is required to know the true identity of the QLr.spa-4B.2 gene.
The SNP marker BobWhite_c1387_798, the most highly associated marker with QLr.spa-5A, the QTL detected from Lillian only, is located on chromosome arm 5AL [42]. To our knowledge there is no designated leaf rust resistance gene reported on this chromosome arm [51]. However, Rosewarne, Singh [56] reported a leaf rust resistance QTL on 5AL in Avocet using a different marker technology making a comparison of relationship difficult. If the QTL in Avocet is different, because no other leaf rust resistance gene has been reported on the long arm of chromosome 5A, QLr.spa-5A would be novel. The consistency of QLr.spa-5A makes it valuable in breeding.
AC Cadillac resistance
AC Cadillac alone contributed two leaf rust resistance QTL, namely QLr.spa-2A.1 and QLr.spa-3B. The QLr.spa-2A.1 mapped in the same region as the QTL that confers multiple disease resistance in Stettler and previously identified in AC Cadillac [22, 24]. Singh, Knox [22] reported the QTL region confers leaf tip necrosis and resistance to leaf rust, stripe rust and powdery mildew. The stripe rust resistance was later confirmed by Bokore, Cuthbert [24]. Like the 2A QTL, the QLr.spa-3B AC Cadillac leaf rust resistance mapped in the region of stripe rust resistance QTL reported in two different studies [22, 24]. The most highly associated SNP marker to QLr.spa-3B, Tdurum_contig79629_538, mapped close to UMN10 [41], Xgwm389 and csSr2 markers that define the region of the slow rusting stem rust gene Sr2 [57, 58]. QLr.spa-3B is in the chromosome region that restricts the development of different fungal diseases with some possibility of relationship to leaf rust resistance gene Lr27 that is known to be associated with stem rust (Sr2) and powdery mildew resistance [59]. Adult plant leaf rust resistance Lr74 is also located close to Sr2 in two US winter wheat cultivars Caldwell [60] and Clark [61]. Further investigation is required to know the relationship of the QLr.spa-3B in Carberry with either Lr27 or Lr74. The multiple disease resistance effects of both QLr.spa-2A.1 and QLr.spa-3B loci make them appealing in breeding.
Lillian resistance
Although Lillian was among the most resistant cultivars in the study, only two QTL (QLr.spa-4A and QLr.spa-7D) segregated from it in the Vesper/Lillian population. Markers for QLr.spa-4A were placed with the SSR markers wmc491 and wmc680 (S1 Fig) that are assigned to chromosome 4AL [34]. Although the seedling resistance gene Lr28 is located on 4AL [51, 62], Lr28 associated SSR markers wmc313, gwm160 and barc78 [63] are about 105 cM distant from the SNP markers defining QLr.spa-4A on our map. The only other resistance reported on 4AL was by Gerard et al. [64] but their QTL is located in a similar region as Lr28/Sr17. None of the four other cultivars we studied harbored QLr.spa-4A making Lillian the exception. Perhaps the gene controlling QLr.spa-4A is less common. Nevertheless it is different from previously reported genes and consequently likely novel. The effectiveness against leaf rust races in Canada and New Zealand of the Lillian QLr.spa-4A allele make it of great interest for resistance breeding.
Stettler resistance
Stettler was the single source of the QLr.spa-2D.2 leaf rust resistance and it possessed a second QTL on 6B, QLr.spa-6B that additionally was detected in Vesper. The QLr.spa-2D.2 locus expressed major resistance that was observed in all tests, which spanned the two countries Canada and New Zealand. The QLr.spa-2D.2 associated SNP markers were assigned to chromosome 2DS [42] as opposed to the Carberry QTL, QLr.spa-2D.1 that is situated on chromosome 2DL based on SNP markers in the QTL interval [42]. on which genes Lr2a linked with gwm484 [65], and Lr22a linked with gwm296, gwm455, and wmc112 [66] are situated. On our consensus map (S1 Fig), the SNP markers flanking QLr.spa-2D.2 place closer to markers for the Lr2a than Lr22a. Stettler likely inherited Lr2a from its immediate parent Superb which is know to possess this gene [44].
Vesper resistance
Three QTL located on chromosomes 1B, 1D and 2A were contributed by Vesper. The 1D, QLr.spa-1D represented a major QTL that consistently expressed in three different populations—Vesper/Lillian, Vesper/Stettler and Carberry/Vesper. The appearance of this QTL in Canada and New Zealand indicated its effectiveness against a diverse spectrum of P. triticina races. RAC875_c2070_566 and BobWhite_c4303_524 that tagged QLr.spa-1D (S1 Fig) resided on 1DS in the same region with the two SSR markers gdm33b and barc149 that are associated with Lr21 [67]. Given Vesper is believed to carry Lr21 through Augusta/Hard White Alpha [68], and QLr.spa-1D mapped to the short arm of chromosome 1D, it is highly likely to be Lr21. Virulence on Lr21 exists in Canada [11]; however, the results of this study suggest that Lr21 was still effective in the northern Great Plains, likely due to the relatively low frequency of races virulent to Lr21 in the inoculum used in this region during the years of testing.
The QTL on 1B (QLr.spa-1B) coincided with a region for stripe rust resistance previously identified on 1BL in Carberry and Vesper [24]. Markers defining the interval of QLr.spa-1B were mapped on chromosome 1BL near SSR markers that are associated with the genomic region considered to be pleiotropic for resistance to multiple fungal diseases, Lr46/Yr29/Pm39/Ltn2 [39, 40]. In addition, wmc44 linked with Lr46 [39, 40] displayed significant association with the leaf rust traits in Vesper/Lillian population that indicated QLr.spa-1B corresponds with Lr46.
Markers associated with the 2A Vesper QTL QLr.spa-2A.2 discovered in Vesper/Stettler and Carberry/Vesper are located on chromosome 2AS [42]. Many genes have been reported on 2AS including Lr17a [69, 70], Lr37/Yr17/Sr38 [71] and Lr65 [72]. However, the relatively short distance observed between the SNP markers associated with QLr.spa-2A.2 and the SSR marker wmc407 associated with Lr17a [69, 70] make QLr.spa-2A.2 the most likely candidate for Lr17a.
QTL common between cultivars
Out of the twenty QTL identified, QTL that are located on chromosomes 6B, 7A, 7B (2 loci), and 7D were detected in two or more wheat cultivars. For example, the QTL on 6B, QLr.spa-6B is in common between Vesper and Stettler. Considering the position of the SNP markers detecting the QTL [42] it is located on the long arm of chromosome 6B. Chromosome 6BS is known for having leaf rust resistance genes Lr36 and Lr53 [73], whereas, 6BL possesses Lr3 [74] and Lr9 [75]. Thus QTL QLr.spa-6B should be different from Lr36 and Lr53 as they are located on different chromosome arms, but it could be Lr3 or Lr9. Virulence to Lr9 has been developing in Canada most likely starting in 2006 reducing the likelihood that QLr.spa-6B is Lr9 [76]. There are no reports of the deployment of Lr3 in the Canadian wheat germplasm making this gene a less likely candidate for QLr.spa-6B. The uniqueness of gene associated with QLr.spa-6B will require further study.
QLr.spa-7A is common between Carberry and Red Fife. The BS00063860_51 SNP marker associated with the Carberry 7A allele and tplb0031i24_1212 with the Red Fife 7A allele are 8.0 cM apart, and located on 7AL [42]. A leaf rust resistance gene that was first identified in the Thatcher-Lr1 near-isogenic Thatcher line RL6003 and temporarily designated as LrCen [77] is the only gene recorded on 7AL. The QLr.spa-7A in the Carberry and Red Fife could be LrCen.
The two QTL on 7B, QLr.spa-7B.1 and QLr.spa-7B.2, are in common between AC Cadillac and Vesper and additionally Red Fife also has QLr.spa-7B.2. Markers associated with the QLr.spa-7B.1 reside on the short arm of chromosome 7B, while markers for the QLr.spa-7B.2 are placed on the long arm of chromosome 7B [42]. No known leaf rust resistance genes are located on the 7BS of bread wheat [51]. However, Herrera-Foessel, Huerta-Espino [78] recently reported the leaf rust resistance gene Lr72 on the 7BS of durum wheat. Interestingly, gwm537 one of the markers associated with Lr72 [78] mapped nearby SNP markers flanking QLr.spa-7B.1 (S1 Fig). In contrast, two genes Lr14a and Lr68 are documented as residing on 7BL [51]. The closest marker to the slow rusting gene Lr68, gwm146 [79] was located between about 3.76 to 26.34 cM from SNP markers tagging QLr.spa-7B.2 in three different populations (S1 Fig). The SSR marker gwm146 is also associated with leaf rust resistance QTL in the durum wheat variety Sachem [80]. Herrera-Foessel, Singh [81] reported the seedling resistance gene Lr14a, tagged with gwm146 and gwm344b, in a Chilean durum cv. Llareta-INIA. Given Lr14a and Lr68 seem to be closely located genes increases the difficulty attributing one of them to QLr.spa-7B.2.
Carberry and Lillian possess QLr.spa-7D that corresponded with the Lr34/Yr18, a region conferring resistance to leaf, stem and yellow rusts, and powdery mildew [82, 83]. The QTL was consistently detected in Carberry and Lillian by SNP12, a diagnostic marker for Lr34 [27]. QLr.spa-7D did not segregate in the Carberry/AC Cadillac as both Carberry and AC Cadillac have Lr34 [84]. The Carberry/AC Cadillac population segregated for 13 QTL; given that Lr34 interacts with other genes to make them more effective [17], it is possible that some of these QTL detected in association with Lr34 may not be detected in other crosses in which Lr34 was not fixed with the resistant allele.
In conclusion, the present study showed that although the spring wheat cultivars Lillian, Carberry, AC Cadillac, Stettler, Vesper and Red Fife may have a few leaf rust resistance loci in common, several QTL differ among cultivars that can be further recombined and deployed as gene stacks. The production of the consensus map integrating SNP and SSR markers, enabled us to understand the similarity between the QTL identified in the present mapping study with those leaf rust resistance genes or QTL previously reported based on SSR marker technology. While many of the identified QTL were previously reported rust resistance genes or QTL, others appear to be novel. For example, the resistance genes identified in Lillian at QLr.spa-4A and Carberry at QLr.spa-5A could be novel genes. In contrast, the 1D QTL in Vesper corresponded with the designated seedling resistance gene Lr21, while the 7D QTL from Lillian and Carberry corresponded with the adult plant resistance gene Lr34. The Carberry resistance QTL QLr.spa-2B.1 corresponded with Lr16, and the 2D QTL from Stettler is more likely Lr2a than Lr22, but could be another unique gene. Carberry demonstrated substantial stacking of genes which could be supplemented with the genes identified in the other adapted varieties with the expectation of increasing efficacy and longevity of resistance to leaf rust with little risk of linkage drag in Canadian wheat breeding programs. Some of the SNP markers associated with the identified QTL have been converted to KASP markers that are being deployed in germplasm evaluation and breeding for marker assisted stacking of leaf rust resistance to develop new varieties.
Supporting information
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Some markers were removed from the linkage maps illustrated here for simplification of the presentation. The test environments are denoted by the test years preceded by an abbreviation of the location as follows: MD, Morden; BD, Brandon; SC, Swift Current, Canada, and LN, Lincoln, New Zealand. The disease phenotypes are abbreviated: LRSEV, leaf rust severity, and LRIR, leaf rust infection response. Chromosome names are followed by population name abbreviations: CCd, Carberry/AC Cadillac; CV, Carberry/Vesper; VL, Vesper/Lillian; VS, Vesper/Stettler; RS, Red Fife/Stettler.
(DOCX)
Acknowledgments
Technical and field support from the Swift Current Research and Development Centre, Morden Research and Development Centre, Brandon Research and Development Centre, Canada, and Plant and Food Research, New Zealand technical staff is greatly appreciated. Genotyping support from the technical staff at the Crop Development Centre Wheat Genomics lab is acknowledged.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Financial support was received from Agriculture and Agri-Food Canada to REK and RDC, the Western Grains Research Foundation to RDC, CJP, and REK, and the Saskatchewan Agriculture Development Fund to CJP, RDC, and REK. Genotyping for the consensus map was supported in part through the “Canadian Triticum Applied Genomics” grant funded by Genome Canada, Genome Prairie, Saskatchewan Ministry of Agriculture, Saskatchewan Wheat Development Commission, Alberta Wheat Development Commission, and the Manitoba Wheat and Barley commission to CJP, RDC, and REK.
References
- 1.McCallum BD, Seto-Goh P, Xue A. Physiologic specialization of Puccinia triticina, the causal agent of wheat leaf rust, in Canada in 2009. Canadian Journal of Plant Pathology. 2013;35(3):338–45. 10.1080/07060661.2013.810669 [DOI] [Google Scholar]
- 2.Kolmer JA. Tracking wheat rust on a continental scale. Current opinion in plant biology. 2005;8(4):441–9. 10.1016/j.pbi.2005.05.001 . [DOI] [PubMed] [Google Scholar]
- 3.McCallum BD, Hiebert CW, Cloutier S, Bakkeren G, Rosa SB, Humphreys DG, et al. A review of wheat leaf rust research and the development of resistant cultivars in Canada. Canadian Journal of Plant Pathology. 2016;38(1):1–18. 10.1080/07060661.2016.1145598 [DOI] [Google Scholar]
- 4.Park RF. Breeding cereals for rust resistance in Australia. Plant Pathology. 2008;57(4):591–602. 10.1111/j.1365-3059.2008.01836.x [DOI] [Google Scholar]
- 5.Bolton MD, Kolmer JA, Garvin DF. Wheat leaf rust caused by Puccinia triticina. Molecular plant pathology. 2008;9(5):563–75. 10.1111/j.1364-3703.2008.00487.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JKM, Hovmøll MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297(5581):537–41. 10.1126/science.1072678 [DOI] [PubMed] [Google Scholar]
- 7.Mcintosh RA, Wellings CR, Park RF. Wheat rusts: An atlas of resistance genes: CSIRO, Australia; 1995. [Google Scholar]
- 8.Line RF, Chen X. Successes in Breeding for and Managing Durable Resistance to Wheat Rusts. Plant Disease. 1995;79:1254–5. [Google Scholar]
- 9.Fetch T, McCallum B, Menzies J, Rashid K, Tenuta A. Rust diseases in Canada. Prairie Soils and Crops Journal. 2011;4:86–96. [Google Scholar]
- 10.Huang L, Brooks SA, Li W, Fellers JP, Trick HN, Gill BS. Map-based cloning of leaf rust resistance gene Lr21 from the large and polyploid genome of bread wheat. Genetics. 2003;164(2):655–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCallum BD, Seto-Goh P, Xue A. The detection of virulence to Lr21 in Canada during 2011. Canadian Journal of Plant Science 2012;92:605. [Google Scholar]
- 12.McCallum BD, Seto-Goh P, Xue A. Physiologic specialization ofPuccinia triticina, the causal agent of wheat leaf rust, in Canada in 2008. Canadian Journal of Plant Pathology. 2011;33(4):541–9. 10.1080/07060661.2011.627950 [DOI] [Google Scholar]
- 13.Spielmeyer W, Mago R, Wellings C, Ayliffe M. Lr67 and Lr34 rust resistance genes have much in common—they confer broad spectrum resistance to multiple pathogens in wheat. BMC Plant Biol. 2013;13:96 10.1186/1471-2229-13-96 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu CG, Friesen TL, Xu SS, Faris JD, Kolmer JA. Identification of novel QTLs for seedling and adult plant leaf rust resistance in a wheat doubled haploid population. Theoretical and applied genetics. 2009;119(2):263–9. 10.1007/s00122-009-1035-0 . [DOI] [PubMed] [Google Scholar]
- 15.Singh RP, Mujeeb-Kazi A, Huerta-Espino J. Lr46: A gene conferring slow-rusting resistance to leaf rust in wheat. Phytopathology. 1998;88(9):890–4. 10.1094/PHYTO.1998.88.9.890 [DOI] [PubMed] [Google Scholar]
- 16.da Silva CGM, Montenegro Stamford TL, Cardoso de Andrade SA, de Souza EL, de Araújo JM. Production of ethanol from mesquite (Prosopis juliflora (SW) D.C.) pods mash by Zymomonas mobilis and Saccharomyces cerevisiae. Electronic Journal of Biotechnology. 2010;13(5):0-. 10.2225/vol13-issue5-fulltext-21 [DOI] [Google Scholar]
- 17.German SE, Kolmer JA. Effect of gene Lr34 in the enhancement of resistance to leaf rust of wheat. Theoretical and Applied Genetics. 1992;84(1–2):97–105. 10.1007/BF00223987 [DOI] [PubMed] [Google Scholar]
- 18.Kolmer JA. A QTL on chromosome 5BL in wheat enhances leaf rust resistance of Lr46. Molecular Breeding. 2015;35(2):74–82. 10.1007/s11032-015-0274-9 [DOI] [Google Scholar]
- 19.Kuchel H, Fox R, Reinheimer J, Mosionek L, Willey N, Bariana H, et al. The successful application of a marker-assisted wheat breeding strategy. Molecular Breeding. 2007;20(4):295–308. 10.1007/s11032-007-9092-z [DOI] [Google Scholar]
- 20.Li Z, Lan C, He Z, Singh RP, Rosewarne GM, Chen X, et al. Overview and Application of QTL for Adult Plant Resistance to Leaf Rust and Powdery Mildew in Wheat. Crop Science. 2014;54(5):1907 10.2135/cropsci2014.02.0162 [DOI] [Google Scholar]
- 21.Da Silva GBP, Zanella CM, Martinelli JA, Chaves MS, Hiebert CW, McCallum BD, et al. Quantitative trait loci conferring leaf rust resistance in hexaploid wheat. Phytopathology. 2018;108(12):1344–54. 10.1094/PHYTO-06-18-0208-RVW [DOI] [PubMed] [Google Scholar]
- 22.Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Campbell HL, et al. Stripe rust and leaf rust resistance QTL mapping, epistatic interactions, and co-localization with stem rust resistance loci in spring wheat evaluated over three continents. Theoretical and applied genetics. 2014;127(11):2465–77. 10.1007/s00122-014-2390-z . [DOI] [PubMed] [Google Scholar]
- 23.Knox RE, Clarke JM, DePauw RM. Dicamba and growth condition effects on doubled haploid production in durum wheat crossed with maize. Plant Breeding. 2000;119(4):289–98. 10.1046/j.1439-0523.2000.00498.x [DOI] [Google Scholar]
- 24.Bokore FE, Cuthbert RD, Knox RE, Randhawa HS, Hiebert CW, DePauw RM, et al. Quantitative trait loci for resistance to stripe rust of wheat revealed using global field nurseries and opportunities for stacking resistance genes. Theoretical and Applied Genetics. 2017;130(12):2617–35. 10.1007/s00122-017-2980-7 [DOI] [PubMed] [Google Scholar]
- 25.McCallum BD, Seto-Goh P, Xue A. Physiologic specialization of Puccinia triticina, the causal agent of wheat leaf rust, in Canada in 2010. Canadian Journal of Plant Pathology. 2016;38(4):440–7. 10.1080/07060661.2016.1261047 [DOI] [Google Scholar]
- 26.Peterson RF, Campbell AB, Hannah AE. A diagramatic scale for estimating rust intensity of leaves and stem of cereals. Canadian Journal of Research. 1948;26:496–500. [Google Scholar]
- 27.Dakouri A, McCallum BD, Walichnowski AZ, Cloutier S. Fine-mapping of the leaf rust Lr34 locus in Triticum aestivum (L.) and characterization of large germplasm collections support the ABC transporter as essential for gene function. Theoretical and applied genetics. 2010;121(2):373–84. 10.1007/s00122-010-1316-7 . [DOI] [PubMed] [Google Scholar]
- 28.Perez-Lara E, Semagn K, Chen H, Iqbal M, N’Diaye A, Kamran A, et al. QTLs asociated with agronomic traits in the Cutler × AC Barrie spring wheat mapping population using single nucleotide polymorphic markers. PLoS ONE 11(8):e0160623 10.1371/journal.pone.0160623 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fowler DB, N’Diaye A, Laudencia-Chingcuanco D, Pozniak CJ. Quantitative trait loci associated with phenological development, low-temperature tolerance, grain quality, and agronomic characters in wheat (Triticum aestivum L.) PLoS ONE 11(3): e0152185 10.1371/journal.pone.0152185 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knox RE, Campbell HL, Clarke FR, Menzies JG, Popovic Z, Procunier JD, et al. Quantitative trait loci for resistance in wheat (Triticum aestivum) to Ustilago tritici. Canadian Journal of Plant Pathology. 2014;36(2):187–201. 10.1080/07060661.2014.905497 [DOI] [Google Scholar]
- 31.Zou J, Semagn K, Iqbal M, Chen H, Asif M, N’Diaye A, et al. QTLs associated with agronomic traits in the Attila × CDC Go spring wheat population evaluated under conventional management. PloS one. 2017;12(2). 10.1371/journal.pone.0171528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, et al. A microsatellite map of wheat. Genetics. 1998;149(4):2007–23. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gupta PK, Varshney RK. The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica. 2000;113(3):163–85. 10.1023/A:1003910819967 [DOI] [Google Scholar]
- 34.Somers DJ, Isaac P, Edwards K. A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theoretical and Applied Genetics. 2004;109(6):1105–14. 10.1007/s00122-004-1740-7 [DOI] [PubMed] [Google Scholar]
- 35.Endelman JB, Plomion C. LPmerge: An R package for merging genetic maps by linear programming. Bioinformatics. 2014;30(11):1623–4. 10.1093/bioinformatics/btu091 [DOI] [PubMed] [Google Scholar]
- 36.Endelman JB. https://cran.r-project.org/web/packages/LPmerge/index.html. 2014.
- 37.Van Ooijen JW. MapQTL® 6: Software for the mapping of quantitative trait loci in experimental populations of diploid species Kyazma BV, Wageningen, The Netherlands: 2009. p. 59 [Google Scholar]
- 38.Voorrips RE. MapChart: Software for the Graphical Presentation of Linkage Maps and QTLs. Journal of Heredity. 2002;93(1):77–8. 10.1093/jhered/93.1.77 [DOI] [PubMed] [Google Scholar]
- 39.Mateos-Hernandez M, Singh RP, Hulbert SH, Bowden RL, Huerta-Espino J, Gill BS, et al. Targeted mapping of ESTs linked to the adult plant resistance gene Lr46 in wheat using synteny with rice. Functional and Integrative Genomics. 2006;6(2):122–31. 10.1007/s10142-005-0017-9 [DOI] [PubMed] [Google Scholar]
- 40.Suenaga K, Singh RP, Huerta-Espino J, William HM. Microsatellite markers for genes lr34/yr18 and other quantitative trait Loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology. 2003;93:881–90. 10.1094/PHYTO.2003.93.7.881 [DOI] [PubMed] [Google Scholar]
- 41.Su Z, Jin S, Zhang D, Bai G. Development and validation of diagnostic markers for Fhb1 region, a major QTL for Fusarium head blight resistance in wheat. Theoretical and Applied Genetics. 2018;131(11):2371–80. 10.1007/s00122-018-3159-6 [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, et al. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant biotechnology journal. 2014;12(6):787–96. 10.1111/pbi.12183 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen W, He Z, Gao F, Liu J, Jin H, Zhai S, et al. A high-density consensus map of common wheat integrating four mapping populations scanned by the 90k SNP array. Frontiers in plant science. 2017;8 10.3389/fpls.2017.01389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCallum BD, Seto-Goh P. The inheritance of leaf rust resistance in the wheat cultivars ’Superb', ’McKenzie’ and ’HY644'. Canadian Journal of Plant Pathology. 2010;32(3):387–95. 10.1080/07060661.2010.499266 [DOI] [Google Scholar]
- 45.Feuillet C, Schachermayr G, Keller B. Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant Journal. 1997;11(1):45–52. 10.1046/j.1365-313x.1997.11010045.x [DOI] [PubMed] [Google Scholar]
- 46.Dyck PL, Kerbere ER. Chromosome locations of three genes for leaf rust resistance in common wheat. Canadian Journal of Genetics and Cytology. 1971;13:480–3. [Google Scholar]
- 47.Feuillet C, Travella S, Stein N, Albar L, Nublat AL, Keller B. Map-based isolation of the leaf rust disease resistance gene Lr10 from the hexaploid wheat (Triticum aestivum L.) genome. PNAS. 2003;100:15253–8. 10.1073/pnas.2435133100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCartney CA, Somers DJ, McCallum BD, Thomas J, Humphreys DG, Menzies JG, et al. Microsatellite tagging of the leaf rust resistance gene Lr16 on wheat chromosome 2BSc. Molecular Breeding. 2005;15(4):329–37. 10.1007/s11032-004-5948-7 [DOI] [Google Scholar]
- 49.Kassa MT, You FM, Hiebert CW, Pozniak CJ, Fobert PR, Sharpe AG, et al. Highly predictive SNP markers for efficient selection of the wheat leaf rust resistance gene Lr16. BMC plant biology. 2017;17(1). 10.1186/s12870-017-0993-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh A, Knox RE, DePauw RM, Singh AK, Cuthbert RD, Campbell HL, et al. Identification and mapping in spring wheat of genetic factors controlling stem rust resistance and the study of their epistatic interactions across multiple environments. Theoretical and Applied Genetics. 2013;126(8):1951–64. 10.1007/s00122-013-2109-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McIntosh RA, Dubcovsky J, Rogers WJ, Morris CF, Appels R, Xia CX. Catalogue of gene symbols for wheat: 2013–14 supplement http://www.shigen.nig.ac.jp/wheat/komugi/genes/symbolClassList.jsp; 2014.
- 52.Oelke LM, Kolmer JA. Genetics of leaf rust resistance in spring wheat cultivars alsen and norm. Phytopathology. 2005;95(7):773–8. 10.1094/PHYTO-95-0773 [DOI] [PubMed] [Google Scholar]
- 53.Schnurbusch T, Paillard S, Schori A, Messmer M, Schachermayr G, Winzeler M, et al. Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theoretical and Applied Genetics. 2004;108(3):477–84. 10.1007/s00122-003-1444-4 [DOI] [PubMed] [Google Scholar]
- 54.Singh S, Bowden RL. Molecular mapping of adult-plant race-specific leaf rust resistance gene Lr12 in bread wheat. Molecular Breeding. 2011;28(2):137–42. 10.1007/s11032-010-9467-4 [DOI] [Google Scholar]
- 55.Bansal UK, Hayden MJ, Venkata BP, Khanna R, Saini RG, Bariana HS. Genetic mapping of adult plant leaf rust resistance genes Lr48 and Lr49 in common wheat. Theoretical and Applied Genetics. 2008;117(3):307–12. 10.1007/s00122-008-0775-6 [DOI] [PubMed] [Google Scholar]
- 56.Rosewarne GM, Singh RP, Huerta-Espino J, Herrera-Foessel SA, Forrest KL, Hayden MJ, et al. Analysis of leaf and stripe rust severities reveals pathotype changes and multiple minor QTLs associated with resistance in an Avocet x Pastor wheat population. Theoretical and applied genetics. 2012;124(7):1283–94. 10.1007/s00122-012-1786-x . [DOI] [PubMed] [Google Scholar]
- 57.Mago R, Simkova H, Brown-Guedira G, Dreisigacker S, Breen J, Jin Y, et al. An accurate DNA marker assay for stem rust resistance gene Sr2 in wheat. Theoretical and applied genetics. 2011;122(4):735–44. 10.1007/s00122-010-1482-7 . [DOI] [PubMed] [Google Scholar]
- 58.Zhang X, Rouse MN, Nava IC, Jin Y, Anderson JA. Development and verification of wheat germplasm containing both Sr2 and Fhb1. Molecular Breeding. 2016;36(7). 10.1007/s11032-016-0502-y [DOI] [Google Scholar]
- 59.Mago R, Tabe L, McIntosh RA, Pretorius Z, Kota R, Paux E, et al. A multiple resistance locus on chromosome arm 3BS in wheat confers resistance to stem rust (Sr2), leaf rust (Lr27) and powdery mildew. Theoretical and applied genetics. 2011;123(4):615–23. 10.1007/s00122-011-1611-y . [DOI] [PubMed] [Google Scholar]
- 60.Kolmer JA, Chao S, Brown-Guedira G, Bansal U, Bariana H. Adult plant leaf rust resistance derived from the soft red winter wheat cultivar ‘caldwell’ maps to chromosome 3BS. Crop Science. 2018;58(1):152–8. 10.2135/cropsci2017.05.0272 [DOI] [Google Scholar]
- 61.Li C, Wang Z, Li C, Bowden R, Bai G, Li C, et al. Mapping of quantitative trait loci for leaf rust resistance in the wheat population ning7840 × clark. Plant Disease. 2017;101(12):1974–9. 10.1094/PDIS-12-16-1743-RE [DOI] [PubMed] [Google Scholar]
- 62.Naik S, Gill KS, Prakasa Rao VS, Gupta VS, Tamhankar SA, Pujar S, et al. Identification of a STS marker linked to the Aegilops speltoides-derived leaf rust resistance gene Lr28 in wheat. Theoretical and Applied Genetics. 1998;97(4):535–40. [Google Scholar]
- 63.Bipinraj A, Honrao B, Prashar M, Bhardwaj S, Rao S, Tamhankar S. Validation and identification of molecular markers linked to the leaf rust resistance gene Lr28 in wheat. Journal of Applied Genetics. 2011;52(2):171–5. 10.1007/s13353-010-0026-9 [DOI] [PubMed] [Google Scholar]
- 64.Gerard GS, Kobiljski B, Lohwasser U, Börner A, Simón MR. Genetic architecture of adult plant resistance to leaf rust in a wheat association mapping panel. Plant Pathology. 2018;67(3):584–94. 10.1111/ppa.12761 [DOI] [Google Scholar]
- 65.Tsilo TJ, Kolmer JA, Anderson JA. Molecular mapping and improvement of leaf rust resistance in wheat breeding lines. Phytopathology. 2014;104(8):865–70. 10.1094/PHYTO-10-13-0276-R [DOI] [PubMed] [Google Scholar]
- 66.Hiebert CW, Thomas JB, Somers DJ, McCallum BD, Fox SL. Microsatellite mapping of adult-plant leaf rust resistance gene Lr22a in wheat. Theoretical and Applied Genetics. 2007;115(6):877–84. 10.1007/s00122-007-0604-3 [DOI] [PubMed] [Google Scholar]
- 67.Hiebert CW, Thomas JB, McCallum BD, Somers DJ. Genetic Mapping of the Wheat Leaf Rust Resistance Gene Lr60 (LrW2). Crop Science. 2008;48:1020–6. [Google Scholar]
- 68.Thomas J, Fox S, McCallum B, Fetch T, Gilbert J, Menzies J, et al. Vesper hard red spring wheat. Canadian Journal of Plant Science. 2013;93:315–21. 10.4141/CJPS2012-233 [DOI] [Google Scholar]
- 69.Bremenkamp-Barrett B, Faris JD, Fellers JP. Molecular mapping of the leaf rust resistance gene Lr17a in wheat. Crop Science. 2008;48(3):1124–8. 10.2135/cropsci2007.07.0379 [DOI] [Google Scholar]
- 70.Zhang JX, Singh RP, Kolmer JA, Huerta-Espino J, Jin Y, Anderson JA. Inheritance of Leaf Rust Resistance in the CIMMYT Wheat Weebill 1. Crop Science. 2008;48(3):1037 10.2135/cropsci2007.08.0455 [DOI] [Google Scholar]
- 71.Helguera M, Khan IA, Kolmer J, Lijavetzky D, Zhong-qi L, Dubcovsky J. PCR assays for the Lr37-Yr17-Sr38 cluster of rust resistance genes and their use to develop isogenic hard red spring wheat lines. Crop Science. 2003;43(5):1839–47. [Google Scholar]
- 72.Mohler V, Singh D, Singrün C, Park RF. Characterization and mapping of Lr65 in spelt wheat ’Altgold Rotkorn'. Plant Breeding. 2012;131(2):252–7. 10.1111/j.1439-0523.2011.01934.x [DOI] [Google Scholar]
- 73.Dadkhodaie NA, Karaoglou H, Wellings CR, Park RF. Mapping genes Lr53 and Yr35 on the short arm of chromosome 6B of common wheat with microsatellite markers and studies of their association with Lr36. Theoretical and Applied Genetics. 2011;122(3):479–87. 10.1007/s00122-010-1462-y [DOI] [PubMed] [Google Scholar]
- 74.Dieguez MJ, Altieri E, Ingala LR, Perera E, Sacco F, Naranjo T. Physical and genetic mapping of amplified fragment length polymorphisms and the leaf rust resistance Lr3 gene on chromosome 6BL of wheat. Theoretical and Applied Genetics. 2006;112(2):251–7. Epub 2005/10/11. 10.1007/s00122-005-0122-0 . [DOI] [PubMed] [Google Scholar]
- 75.Schachermayr G, Siedler H, Gale MD, Winzeler H, Winzeler M, Keller B. Identification and localization of molecular markers linked to the Lr9 leaf rust resistance gene of wheat. Theoretical and Applied Genetics. 1994;88(1):110–5. 10.1007/BF00222402 [DOI] [PubMed] [Google Scholar]
- 76.McCallum BD, Seto-Goh P. Physiologic specialization of Puccinia triticina, the causal agent of wheat leaf rust, in Canada in 2006. Canadian Journal of Plant Pathology. 2009;31(1):80–7. 10.1080/07060660909507575 [DOI] [Google Scholar]
- 77.McCallum B, Hiebert CW. Characterization of the Wheat Leaf Rust Resistance Gene LrCen Poster abstract, Pant and Animal Genome XX, January 14–18, 2012, San Diego, CA. 2012. [Google Scholar]
- 78.Herrera-Foessel SA, Huerta-Espino J, Calvo-Salazar V, Lan CX, Singh RP. Lr72 confers resistance to leaf rust in durum wheat cultivar atil C2000. Plant Disease. 2014;98(5):631–5. 10.1094/PDIS-07-13-0741-RE [DOI] [PubMed] [Google Scholar]
- 79.Herrera-Foessel SA, Singh RP, Huerta-Espino J, Rosewarne GM, Periyannan SK, Viccars L, et al. Lr68: a new gene conferring slow rusting resistance to leaf rust in wheat. Theoretical and Applied Genetics k. 2012;124(8):1475–86. 10.1007/s00122-012-1802-1 . [DOI] [PubMed] [Google Scholar]
- 80.Singh A, Pandey MP, Singh AK, Knox RE, Ammar K, Clarke JM, et al. Identification and mapping of leaf, stem and stripe rust resistance quantitative trait loci and their interactions in durum wheat. Molecular breeding: new strategies in plant improvement. 2013;31(2):405–18. 10.1007/s11032-012-9798-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herrera-Foessel SA, Singh RP, Huerta-Espino J, William HM, Garcia V, Djurle A, et al. Identification and molecular characterization of leaf rust resistance gene Lr14a in durum wheat. Plant Disease. 2008;92(3):469–73. 10.1094/PDIS-92-3-0469 [DOI] [PubMed] [Google Scholar]
- 82.Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, et al. Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theoretical and Applied Genetics. 2009;119(5):889–98. 10.1007/s00122-009-1097-z . [DOI] [PubMed] [Google Scholar]
- 83.McCallum BD, Humphreys DG, Somers DJ, Dakouri A, Cloutier S. Allelic variation for the rust resistance gene Lr34/Yr18 in Canadian wheat cultivars. Euphytica. 2012;183(2):261–74. 10.1007/s10681-011-0519-6 [DOI] [Google Scholar]
- 84.Hiebert CW, Fetch TG, Zegeye T, Thomas JB, Somers DJ, Humphreys DG, et al. Genetics and mapping of seedling resistance to Ug99 stem rust in Canadian wheat cultivars ’Peace’ and ’AC Cadillac'. Theoretical and Applied Genetics. 2011;122(1):143–9. 10.1007/s00122-010-1430-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Some markers were removed from the linkage maps illustrated here for simplification of the presentation. The test environments are denoted by the test years preceded by an abbreviation of the location as follows: MD, Morden; BD, Brandon; SC, Swift Current, Canada, and LN, Lincoln, New Zealand. The disease phenotypes are abbreviated: LRSEV, leaf rust severity, and LRIR, leaf rust infection response. Chromosome names are followed by population name abbreviations: CCd, Carberry/AC Cadillac; CV, Carberry/Vesper; VL, Vesper/Lillian; VS, Vesper/Stettler; RS, Red Fife/Stettler.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.


