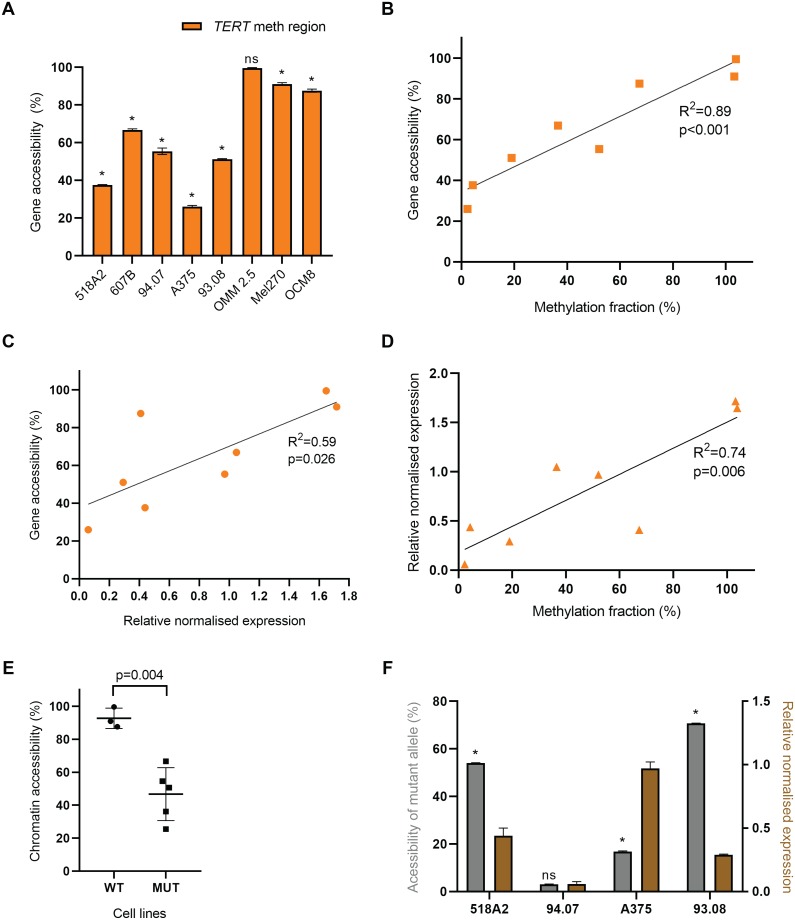
Fig 6. Accessibility of TERTp around cg11625005 in 8 melanoma cell lines.
Cell lines (518A2, 607B, 94.07, A375, 93.08, OMM2.5, Mel270 and OCM8) were analysed with the EpiQ chromatin kit, and ddPCR was performed using primers and probes for positive control gene GAPDH and for the TERT methylation region, a 231-bp amplicon around cg11625005. Accessibility (%) was calculated by the ratio of the digested sample to its matched undigested sample, subtracted from 1, and subsequently normalised against the positive control GAPDH. A. Accessibility of the TERT methylation region relative to GAPDH (mean ± SD, multiple t-tests, one t-test per cell line, *p<0.001, ns p = 0.149). B & C. Correlation plots of gene accessibility around cg11625005 with the MF (%) of cg11625005 obtained through ddPCR (B), or with normalised expression levels via qPCR (C). Linear regression and correlation analysis were performed (F(1.6) = 49.9, r = 0.95, p<0.001 and F(1,6) = 8.6, r = 0.77, p<0.05, respectively). D. Correlation plot between MF (%) of cg11625005 obtained through ddPCR and normalised expression levels via qPCR. Linear regression and correlation analysis were performed (F(1.6) = 16.92, r = 0.86, p<0.05). E. Comparison of WT (OMM2.5, Mel270 and OCM8) and mutated (518A2, 607B, 94.07, A375, 93.08) TERT-expressing cell lines subsets regarding chromatin accessibility (two-tailed unpaired t-test; t = 4.63, df = 6; p<0.005). F. Accessibility of mutant allele (%) in a subset of 4 TERTp-mutated cutaneous cell lines (518A2, 94.07, A375 and 93.08) calculated as described in Material and Methods (mean ± SD, multiple t-tests, one t-test per cell line, ns p = 0.171; *p<0.001) and the TERT mRNA expression in the respective cell lines (mean ± SEM).