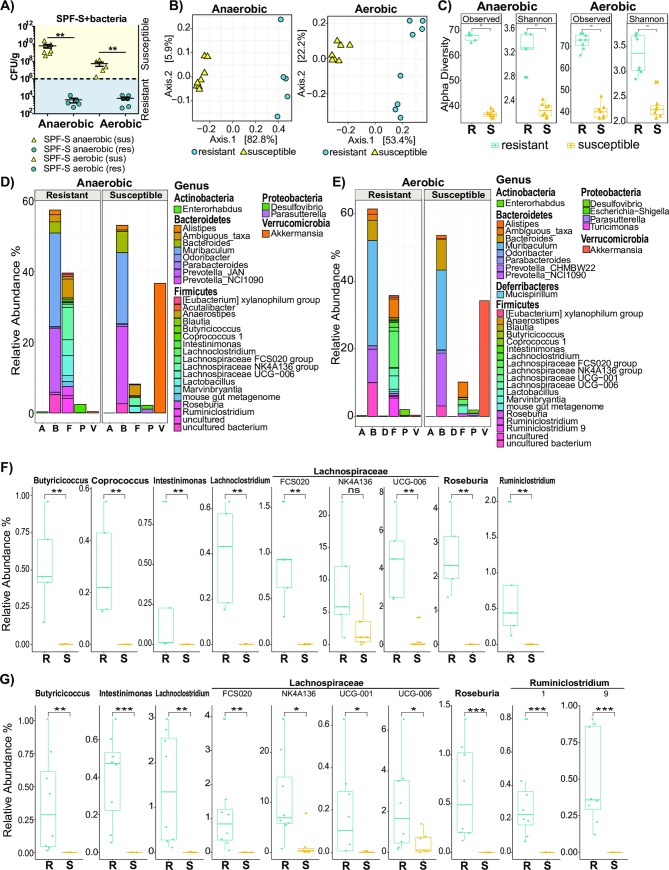
Fig 5. Resistance of SPF-S mice is associated with higher diversity and abundance of bacteria in the Firmicutes phylum.
(A) Assignment of SPF-S mice receiving a cultivated fraction of anaerobically or aerobically cultivated bacteria from the SPF-R mice to the class “resistant” and “susceptible” according to the cecal CFUs of C. rodentium after day 3 p.i.. A threshold of 106 was used for discrimination of both groups. (B) Fecal microbiota of resistant and susceptible SPF-S mice of the “anaerobically treated” and “aerobically treated” group was analyzed using 16S rRNA gene sequencing after cohousing using a principal coordinates analysis (PCoA) plot (C) α-diversity was determined using Chao1 and Shannon index. P values indicated represent a non-parametric Wilcoxon signed rank test *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (D-E) Fecal microbiota of resistant and susceptible “anaerobically treated” and “aerobically treated” SPF-S mice were analyzed using 16S rRNA gene sequencing. Relative abundances of bacterial genera are shown and grouped according to their phylum. Bars represent the mean of all mice within the group. Representative data derived from three independent experiments are shown. (F-G) Relative abundance of significantly different SCFA producing members of treated SPF-S mice of the genera Butyricicoccus, Coprococccus, Intestinimonas, Lachnoclostridium, Lachnospiraceae, Roseburia and Ruminoclostridium are shown. P values indicated represent a non-parametric Wilcoxon signed rank test *p<0.05, **p<0.01, ***p<0.001.