Dear Editor,
Previous reports indicated that the emergence of the novel coronavirus (SARS-CoV-2) infection (COVID-19) had raised global concern and was characterized as a pandemic event by the World Health Organization on March 11, 2020.1, 2, 3 Till March 18, 2020, it has spread to 146 countries, including Taiwan.4 People in Taiwan and mainland China travel frequently, which put Taiwan at a great risk of acquiring an epidemic of COVID-19. Taiwan has been on constant alert and react rapidly to epidemics change from China ever since the severe acute respiratory syndrome (SARS) epidemic in 2003 and has done much effort on the containment of COVID-19 with success.5 Till March 18, 2020, there are only 100 cases of COVID-19 noted in Taiwan, including 79 imported cases and 21 cases belonging to seven occasions of limited local transmission (six family clusters and two transmissions in social societies).4
To contain the epidemics of COVID-19, prevention from both import and export of contagious people is an essential intervention. It is also important to clearly clarify the infection source in order to initiate an efficient and successful contact tracing for the SARS-CoV-2 infected patients, and thereafter people exposed to the contagious patients could be quarantined to avoid further disease spreading. Here we present a COVID-19 patient whose infection source could not be completely clarified initially, and later was illuminated by using the phylogenetic analysis of the isolated virus.
This 66-year-old Taiwanese woman was well before. She traveled to Dubai from January 29 to February 10, 2020, and Egypt from February 11 to February 21, 2020. When she stayed in Egypt, she ever participated an eight-day tourism on a Nile cruise boat. She returned to Taiwan via an international airline on February 21, 2020. She began to suffer from general malaise, myalgia, cold sweating, productive cough, and sore throat since February 18, 2020. She reported that there were another 16 persons in the same tourism group had similar symptoms at that time. Her symptoms persisted despite medication, prescribed on February 21 by a local medical doctor. On February. 26, the cough exacerbated, and she developed chest tightness, abdominal upset and vomiting. She visited the Department of Emergency of a teaching hospital in Taipei on February 28. No fever was noted during her disease course. In the context of her travel history and prominent respiratory symptoms, a nasopharyngeal swab was taken for test of SARS-CoV-2 by real-time reverse transcription-polymerase chain reaction (RT-PCR), and the result was positive. She was then transferred to a negative-pressure isolation room as a case of COVID-19.
The most interesting point of this patient is where she contracted her COVID-19. By history, she is more likely to contract SARS-CoV-2 infection while travelling abroad. However, despite that the median incubation period of COVID-19 was 5.1 days, it might be as long as more than three weeks in some extreme cases.6 , 7 Therefore, an argument that she got the infection while she was in Taiwan couldn't be excluded completely.
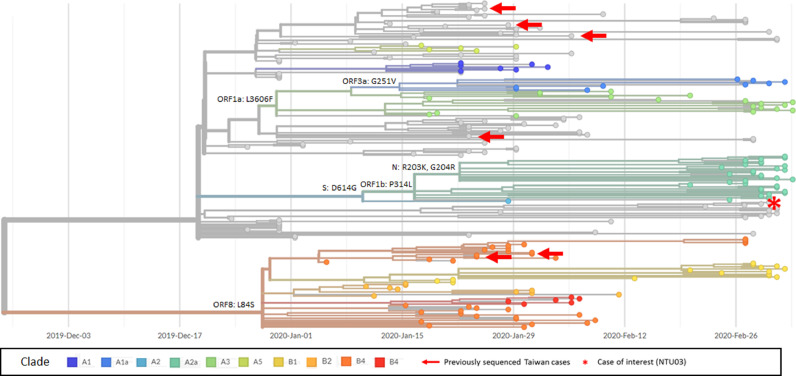
To clarify this argument, more virologic studies were conducted. Virus whole genome sequencing was conducted for the SARS-CoV-2 isolate (NTU03) from her throat swab collected on March 2, 2020. The derived NTU03 sequence was most similar to clade A2a with only 5 nucleotide differences, which included 2 synonymous mutations (Orf 1b/5410 CTA>TTA, and orf 3a/819 GTG>GTT), 2 nonsynonymous mutations (Orf 1b/799 G>V, and orf 3a/57Q>H), and a mutation within 5′UTR (Table 1). An average of 12 nucleotide differences were observed between NTU03 and sequences of other clades, whereas an average of 10 nucleotide differences were observed with other previous viruses isolated from Taiwan. The phylogenetic analysis also reveals that the NTU03 belongs to clade A2a (Fig. 1 ).
Table 1.
Summary of SARS-CoV-2 alignment.
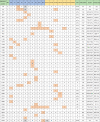 |
SARS-CoV-2 genetic sequences, including NTU03, all 6 previously submitted Taiwan sequences, and representative sequences for clades A1, A1a, A2, A2a, A3, A5, B1, B2, and B4, were aligned and compared. MN908947 was used as the reference sequence, and orange shaded nucleotides indicated nucleotides different to the reference nucleotide, and gray shaded positions indicated lack of nucleotide information. The NTU01 and CDC03 are derived from the Case 3 patient, and CDC04 and CGMH1 are from the from the Case 4 patient. Sequence data were obtained from GISAID (https://www.gisaid.org/CoV2020/) . The GISAID accession number for each representative clade sequence: A1a, hCoV-19/Switzerland/1,000,477,102/2020|EPI_ISL_413,019; A1, hCoV-19/Singapore/11/2020|EPI_ISL_410,719; A2a, hCoV-19/Italy/UniSR1/2020|EPI_ISL_413,489; A2, hCoV-19/Germany/BavPat1/2020|EPI_ISL_406,862; A3, hCoV-19/Australia/NSW13/2020|EPI_ISL_413,599; A5, hCoV-19/USA/CA5/2020|EPI_ISL_408,010; B1, hCoV-19/USA/WA12-UW8/2020|EPI_ISL_413,563; B2, hCoV-19/USA/TX1/2020|EPI_ISL_411,956; B4-1, hCoV-19/South Korea/KUMC03/2020|EPI_ISL_413,513; B4-2, hCoV-19/USA/CA7/2020|EPI_ISL_411,954.
Fig. 1.
Phylogenetic analysis of the full-length SARS-CoV-2 sequences. The phylogeny tree analysis was conducted to determine the clade of NTU03 (red asterisk) and its relationship to other viral sequences derived from case patients identified in Taiwan (red arrows). The phylogenetic tree was generated and modified for display purposes from Nextstrain (https://nextstrain.org/ncov),9 which uses genetic sequences and metadata from GISAID (https://www.gisaid.org/CoV2020/) and sequence submission date for the horizontal axis.10 The phylogenetic tree was generated at 2020/03/09 6PM (GMT+8) with a total of 240 viral genomes sampled (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
Based on the results of whole genome sequencing and phylogenetic analysis, NTU03 belongs to clade A2a, in which all other of the reported case patients were currently either from Europe or travelled to Europe recently according to the information provided by the laboratories who submitted the clade A2a sequences to the Global Initiative on Sharing All Influenza Data (GISAID). None of the previously submitted sequences of viruses isolated from Taiwan were assigned to clade A2a or A2.
With the limited transmission clusters in Taiwan and the fact that NTU03 exhibits at least 8 unique nucleotide difference compared to other previously reported viruses from Taiwan, we conclude that it is much more probable that the present patient was infected during her travelling abroad. To our best knowledge, in late February and early March, several foreign COVID-19 patients who had travel histories to Egypt were reported. Further phylogenetic analysis including viral sequences derived from the 45 confirmed SARS-CoV-2 infections on the quarantined Nile cruise boat will help to delineate the outbreak on Nile tourism boat and its impact on the COVID-19 epidemic.8
Declaration of Competing Interest
None to declare.
Acknowledgments
Acknowledgments
We thank all the persons involved in the response to this outbreak.
Financial support
This study was financially supported by grant from the “Center of Precision Medicine” from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
References
- 1.Tang J.W., Tambyah P.A., Hui D.S.C. Emergence of a novel coronavirus causing respiratory illness from Wuhan, China. J Infect. 2020;80(3):350–371. doi: 10.1016/j.jinf.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Z., Xiao K., Zhang X., Roy A., Shen Y. Emergence of SARS-like coronavirus in China: an update. J Infect. 2020 doi: 10.1016/j.jinf.2020.03.010. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization[Internet]. Statement on the second meeting of the International Health Regulations (2005) Emergency Committee regarding the outbreak of novel coronavirus (2019-nCoV). [cited 2020 Mar 18]. Available from:https://www.who.int/emergencies/diseases/novel-coronavirus-2019/events-as-they-happen.
- 4.Taiwan Centers for Disease Control. Situation of COVID-19 in Taiwan. [cited 2020 Mar 18]. Available from:https://sites.google.com/cdc.gov.tw/2019ncov/taiwan.
- 5.Wang C.J., Ng C.Y., Brook R.H. Response to COVID-19 in Taiwan: big data analytics, new technology, and proactive testing. JAMA. 2020 doi: 10.1001/jama.2020.3151. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 6.Backer J.A., Klinkenberg D., Wallinga J. Incubation period of 2019 novel coronavirus (2019-nCoV) infections among travelers from Wuhan, China. Euro Surveill. 2020;25(5) doi: 10.2807/1560-7917.ES.2020.25.5.2000062. 20028 January 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med. 2020 doi: 10.7326/M20-0504. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Washington Post. Egypt tires to reassure tourists as coronarivus spikes. [cited 2020 Mar 18]. Available from:https://www.washingtonpost.com/world/middle_east/egypt-tries-to-reassure-tourists-as-coronavirus-spikes/2020/03/08/ab1dfbf6-6160-11ea-8a8e-5c5336b32760_story.html.
- 9.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34(23):4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagulenko P., Puller V., Neher R.A. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4(1) doi: 10.1093/ve/vex042. vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]