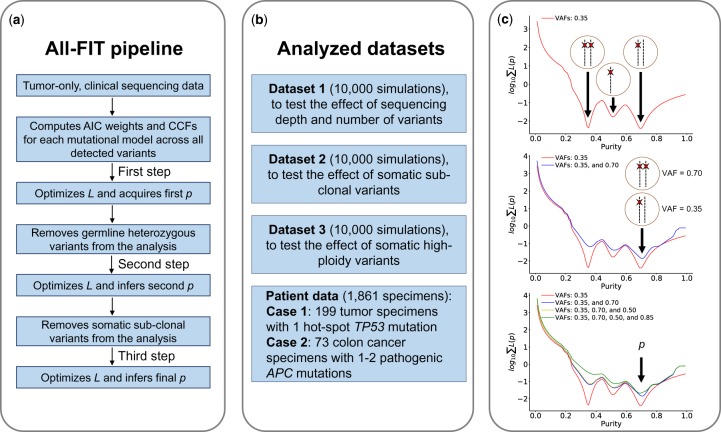
Fig. 2.
Schematic view of All-FIT’s implementation and the analyzed datasets. (a) All-FIT follows three steps for estimating p, first assuming all variants are clonal, then removing germline heterozygous variants, and finally excluding sub-clonal somatic variants. (b) Three simulated datasets are generated to test the effect of sequencing depth and number of variants, the impacts of sub-clonal somatic variants as well as high-ploidy somatic variants. We also apply All-FIT to 1861 patient specimens, particularly 199 tumor specimens harboring hot-spot TP53 mutations and 73 colon cancer specimens harboring pathogenic APC mutations. (c) If only a group of variants with observed VAFs of 0.35 exists in a specimen, these variants cannot be distinguished between three different somatic mutational models of heterozygous, under LOH, or under copy-neutral LOH, without knowing specimen’s tumor content. If a second group of variants is detected with observed VAFs of 0.70, p = 0.70 can classify variants with observed VAFs of 0.35 and 0.70 as heterozygous and copy-neutral LOH somatic mutations, respectively. Detection of additional variants with VAFs of 0.50 and 0.85 improves confidence in estimating purity