Fig. 5.
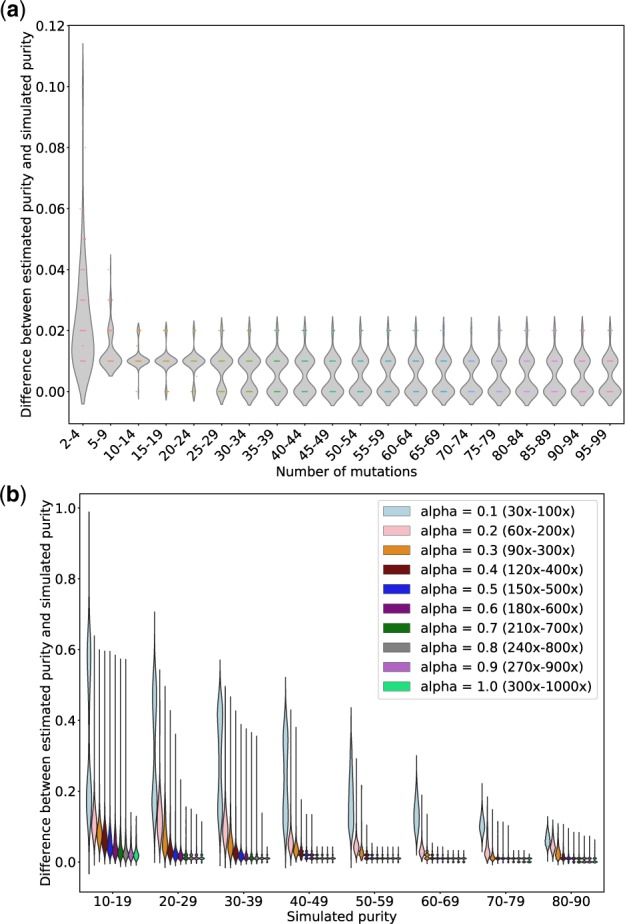
The impact of number of mutations and sequencing depth on All-FIT’s accuracy. (a) Using simulations from Dataset 1, 100 samples are chosen, and each is subsampled repeatedly into 100 permuted sets with varying number of variants ranging from 2 to 99. The x-axis represents the number of variants, and the y-axis represents the median difference between estimated and simulated purities across permutations for each sample (without considering the purity CIs). These results show that the accuracy of All-FIT is independent of the number of variants, when there are at least 10 variants per sample. (b) Using simulations from Dataset 1, 10000 simulated sets are regenerated by changing the variants’ total depth with factors ranging from 0.1 to 1.0; we repeat this permutated subsampling 100 times for each simulated sample at all values of alpha. The x-axis represents the simulated purity (with 1100–1300 samples for each purity categories), and the y-axis represents the median difference between estimated and simulated purities across permutations for each sample. These results show that All-FIT’s accuracy is dependent on the sequencing depth given specimen purity
