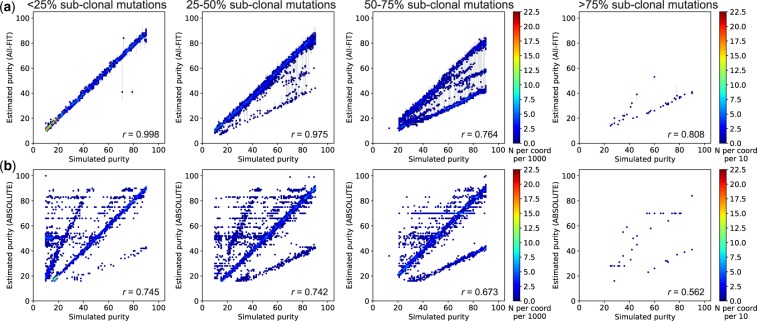
Fig. 6.
Presence of sub-clonal mutations reduces the correlation coefficient between simulated purity and estimated purity using Dataset 2. (a) All-FIT accurately imputes p in simulated sets when percentage of sub-clonal mutations is <50%, beyond which, it increasingly underestimates purity at one-half of the simulated value. (b) ABSOLUTE often fails to predict p correctly even when the percentage of sub-clonal mutations is <25%. The scale-bar represents the number of overlapping samples at each coordinate per 1000 for first three panels from the right, and per 10 for the leftmost panel. All-FIT shows high correlation when <50% of variants are sub-clonal, but overall, its correlation is relatively higher than those of ABOSLUTE, regardless of the percentage of sub-clonal mutation (Supplementary Fig. S1)