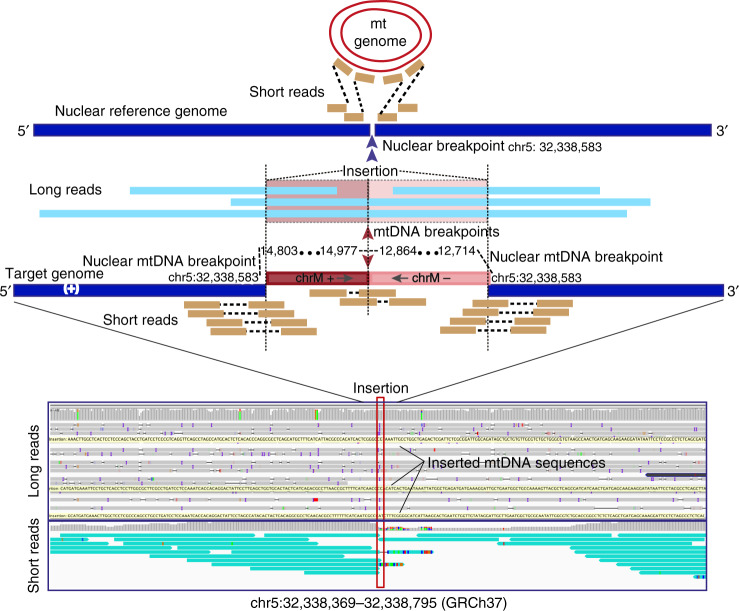
Fig. 6. Validation of NUMTs using long-read (Oxford Nanopore PromethION) sequencing.
Example shown is a concatenated mtDNA NUMT detected by both long-read sequencing and short-read sequencing. mtDNA and nuclear genome reference sequences are shown on the top in red and navy. Aligned short reads shown in orange and long reads in light blue. The sequenced genome included concatenated mtDNA sequences (fragment 1: mt 14803-14977 (+) and fragment 2: 12864-12714 (−)) inserted into chr5: 32,338,583. The two mtDNA fragments are coloured in red and plink. The sequences of the long reads aligned to two mtDNA fragments are also highlighted in red and pink. The remaining sequences aligned to nuclear genome in the same region. The IGV screenshot shows both long-read (top) and short-read (bottom) alignments. The insertion point on the nuclear genome is highlighted by the red box. The yellow bars on the long-read alignment are the large insertions from mtDNA sequences. Teal bars on the short-reads alignment are the discordant reads which one read aligned to nuclear genome and their mate reads aligned to mtDNA genome.