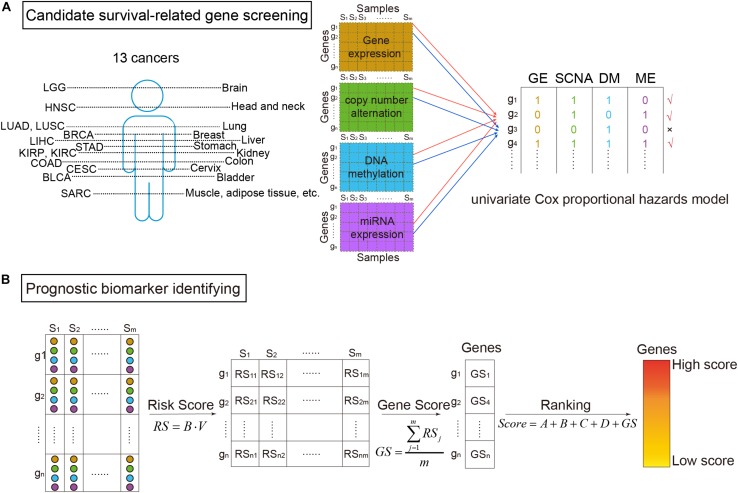
FIGURE 1.
The workflow of survival-related genes identification. (A) Candidate survival-related gene screening. DNA methylation, gene expression, somatic copy-number alteration, and microRNA (miRNA) expression profiles of TCGA for the same samples were extracted. miRNA expression data were corresponding to genes according to miRNA–mRNA interactions. Then, we got four types of data in the same samples and the same genes. On each omics data, univariate Cox proportional hazards model was utilized to identify survival-related genes. Only the genes associated with survival in more than two types of data were considered to be candidate genes. (B) Prognostic biomarker identifying. For the selected candidate genes, the multivariate Cox proportional hazards model was then applied to get risk scores (RS). Further, scores for ranking genes were obtained by calculating GS scores. In which, A, B, C, and D were binary variables indicating whether the gene was survival-related at the four omics data or not (“1” for related and “0” for not), respectively. The high ranked genes were identified survival-related.