Table 1.
Computational predictions of interactions between ligands and MTase domain of human DNMT1 by molecular docking. Concordant amino acid residues involved in the predicted interactions are indicated in bold.
| Ligand | CID | 2D Molecular Structures * |
Binding Energy (Kcal/mol) |
Max RMSD ** | Hydrophobic Contacts | Hydrogen Bonds |
|---|---|---|---|---|---|---|
|
S-adenosyl-homocysteine (SAH) |
439155 |
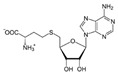
|
−8.3 | 4.547 |
Phe1145, Leu1151, Glu1168, Cys1191, Leu1247, Ala1579, Val1580 |
Met1169, Asp1190, Ans1578 |
| caffeic acid | 689043 |
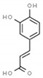
|
−6.7 | 6.809 | Ser1146, Gly1147, Cys1148, Asn1578, Ala1579 | Gly1149, Gly1150, Leu1151, Val1580 |
| hydrocinnamic acid |
107 |
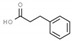
|
−5.2 | 7.170 | Ser1146, Gly1147, Asn1578, Gly1223 | Leu1151, Val1580 |
| p-coumaric acid | 637542 |
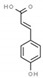
|
−6.0 | 6.049 | Gly1147, Asn1578, Ala1579 | Gly1149, Gly1150, Leu1151, Val1580 |
| (−)-epigallocatchin−3-gallate | 65064 |
|
−10.4 | 0.1564 | Arg1312, Asn1578, Val1580, Gly1223, Gly1147, Phe1145 |
Glu1266, Arg1310 |
CID–PubChem Compound ID number; (*) Retrieved from ChemSpider (https://www.chemspider.com/Default.aspx); (**) RMSD-Root-Mean-Square Deviation.
