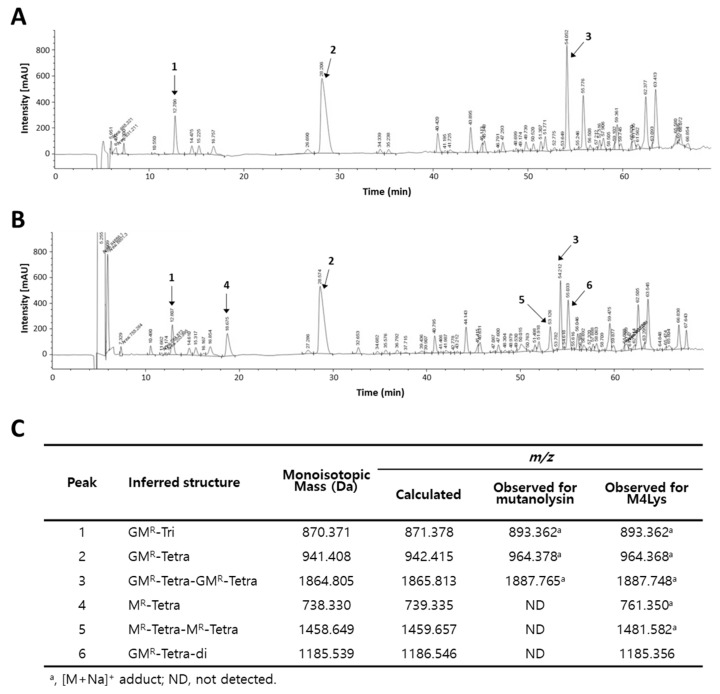
Figure 4.
Determination of M4Lys cleavage sites. Analysis of E. coli MG1655 peptidoglycan digested by (A) mutanolysin and (B) mutanolysin with purified M4LysΔTMD. Soluble muropeptides were reduced and analyzed by RP-HPLC coupled to MS. Peaks corresponding to m/z values matching previously identified muropeptides are numbered. The fragmentation pattern (peaks 1, 2, and 3) is typical of Tri (L-Ala-D-Glu-m-DAP), Tetra (L-Ala-D-Glu-m-DAP-D-Ala), and Tetra-Tetra muropeptides, respectively. The fragmentation event leading to the loss of a nonreduced GlcNAc residue (203.078, theoretical mass; peaks 4 and 5) indicates the N-acetylmuramidase activity of M4LysΔTMD. Cleavage of D-Glu-mDAP crosslink (peak 6) shows the endopeptidase activity of M4LysΔTMD; (C) Inferred structures, theoretical monoisotopic masses, and theoretical and observed m/z values of individual peaks are tabulated. MR, reduced MurNAc; G, GlcNAc; Di, m-DAP (meso-diaminopimelic acid)-D-Ala; Tri, L-Ala-D-Glu-m-DAP; and Tetra, L-Ala-D-Glu-m-DAP-D-Ala.