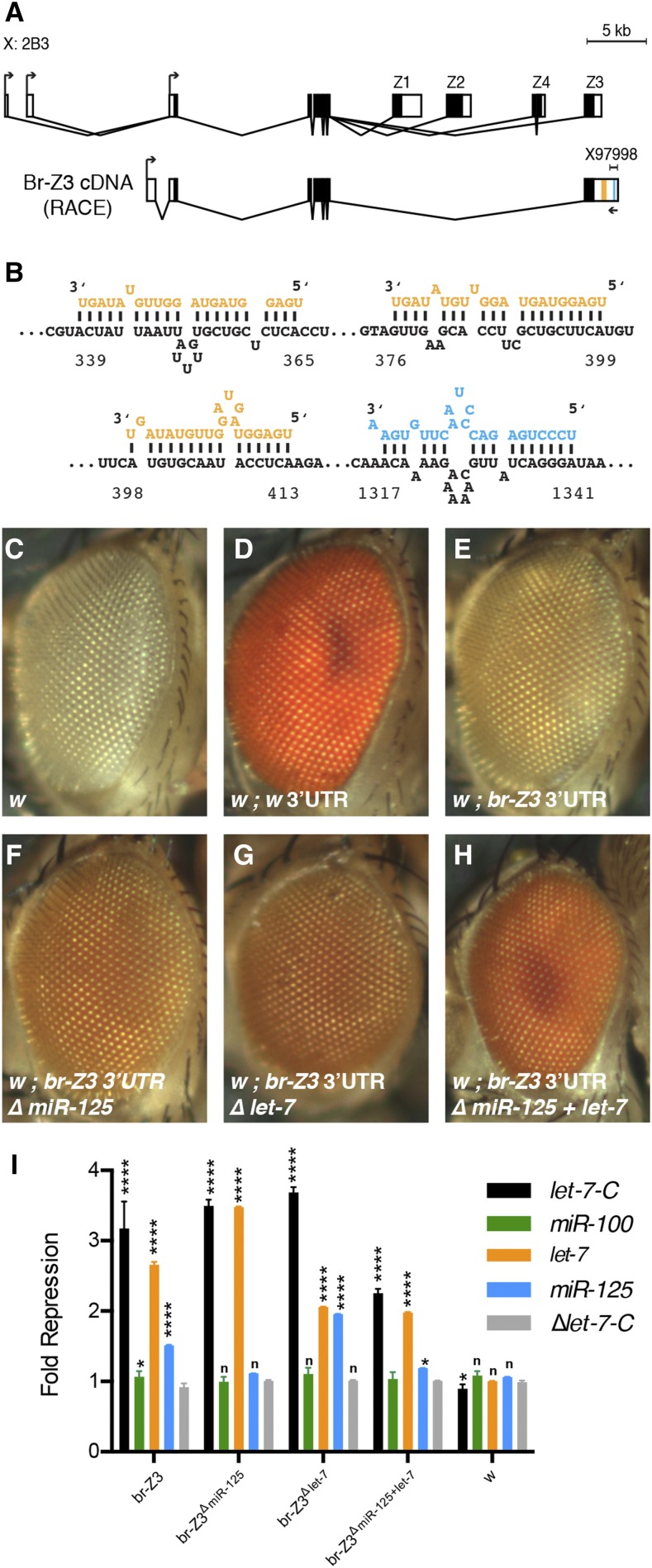
Figure 1.
broad-Z3 3′UTR contains functional let-7 and miR-125 binding sites. (A) Organization of the broad locus (top) and RACE-identified br-Z3 transcript (bottom). Locations of Z1-Z4 encoding exons and predicted let-7 and miR-125 binding sites (orange and blue lines, respectively) are shown. X97998 indicates the location of a partial broad cDNA predicted to contain a miR-125 binding site originally reported in Makunin et al. 1996. (B) Sequences of predicted let-7-C binding sites in br-Z3. let-7, miR-125, and br-Z3 3′UTR sequences are shown in orange, blue, and black, respectively, and the br-Z3 3′UTR sequence is numbered relative to the stop codon. (C-H) Eyes of white mutant flies harboring reporter transgenes containing a white (D), br-Z3 (E), or mutated br-Z3 3′UTR (F-H). (I) Fold repression of luciferase reporters containing full-length wildtype br-Z3 3'UTR, mutated br-Z3 3′UTR, or white 3′UTR in cell cultures in which let-7-C miRNAs were ectopically expressed either together or individually. ****, P < 0.0001; *P < 0.05; n.s. = not significant. Error bars = ± SD.