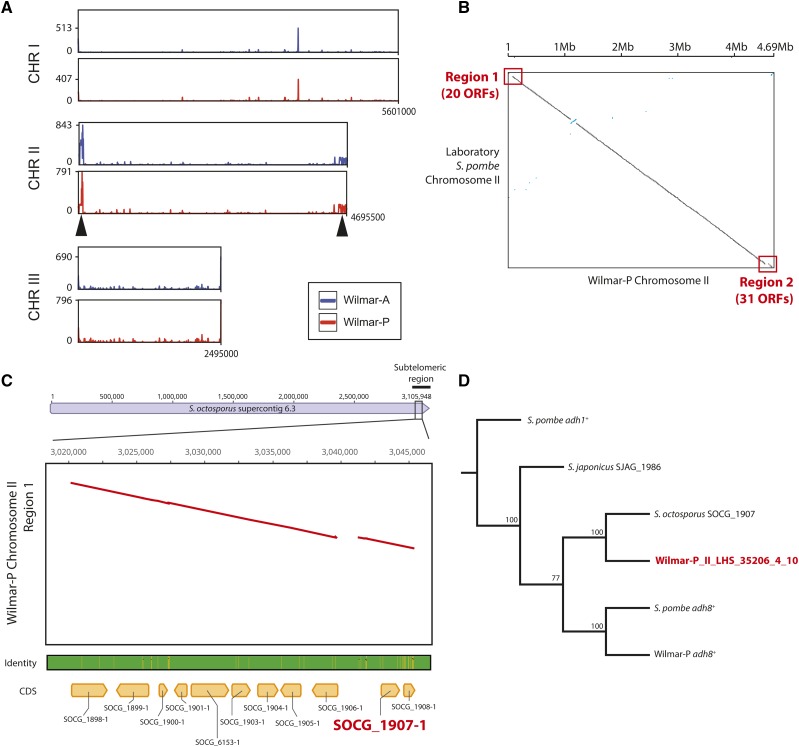
Figure 5.
The Wilmar-P genome contains Schizosaccharomyces octosporus genes. A) Coverage plots of Wilmar-A and Wilmar-P Illumina reads that did not align to the laboratory S. pombe genome mapped back to the de novo assembled Wilmar-P genome. Average read depth calculated using a 10kb sliding window with a 1kb step size. Y-axis indicates depth of coverage and is capped at 900 (Wilmar-A) or 800 (Wilmar-P). X-axis shows position within each chromosome with the total length of each in the Wilmar-P genome. B) Whole chromosome alignment of Wilmar-P chromosome II vs. laboratory S. pombe chromosome II showing two regions unique to the Wilmar-P genome and the number of open reading frames (ORFs) detected within each. C) Whole chromosome alignment of S. octosporus supercontig 6.3 vs. Wilmar-P chromosome II Region 1. Map is centered on the region showing conserved synteny. Orthologous S. octosporus genes are labeled. The gene labeled in red, SOCG-1907-1, encodes an alcohol dehydrogenase orthologous to S. pombe adh8+ used in (D) to construct a neighbor joining tree. D) Tree generated using 100 bootstrap replicates with laboratory S. pombe adh1+ as the outgroup.