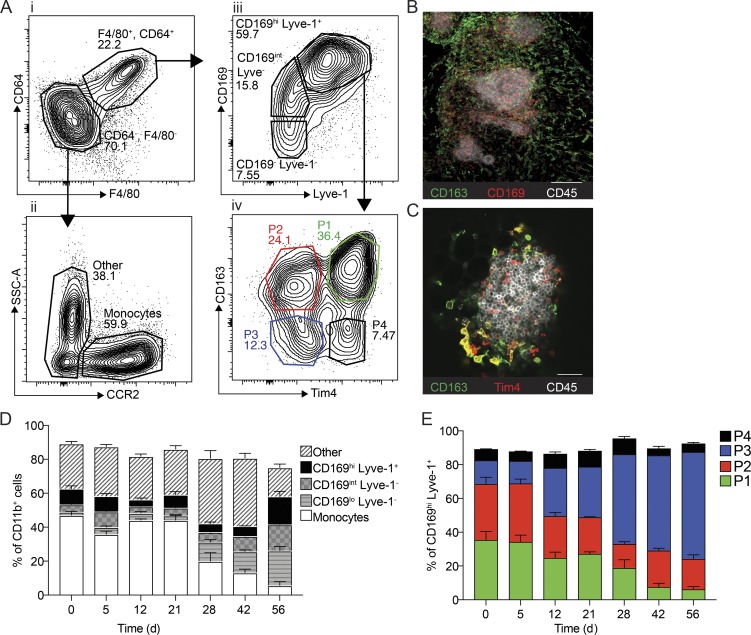
Figure 3.
Characterization of macrophage subsets in omentum by flow cytometry and confocal microscopy. (A) Flow cytometry analysis of the myeloid cell compartment in omentum. Myeloid cells are gated as (i) Live, CD45.2+, singlets, Linneg (CD5, CD19, NK1.1, Ly6G, Siglec F), CD11b+, and subsequently macrophages were gated as F4/80+, CD64+. Monocytes were gated as (ii) F4/80−, CD64−, CCR2+. F4/80−, CD64−, and CCR2− CD11b+ myeloid cells were designated as other. F4/80+ CD64+ macrophages were further gated based on (iii) CD169 and Lyve-1 expression, and finally CD169hi Lyve-1+ cells were further divided into four subpopulations based on (iv) CD163 and Tim4 expression. SSC-A, side scatter area. (B) Whole-mount imaging of FALCs (CD45, white; CD169+, red; CD163+, green) in the omentum; scale bar: 100 µm. (C) Whole-mount imaging of FALCs (CD45, white; CD163+, green; Tim4+, red) in omentum; scale bar: 50 µm. (D) Flow cytometry analysis and relative distribution of CD11b+ myeloid cell populations shown in A, i–iii during tumor growth. (E) Flow cytometry analysis and relative distribution of CD169hi Lyve-1+ macrophage subsets shown in A, iv during tumor growth. Whole-mount confocal imaging analysis is representative of n = 4 in two independent experiments. Flow cytometric data are represented as mean ± SEM of n = 4 and are representative of two independent experiments.