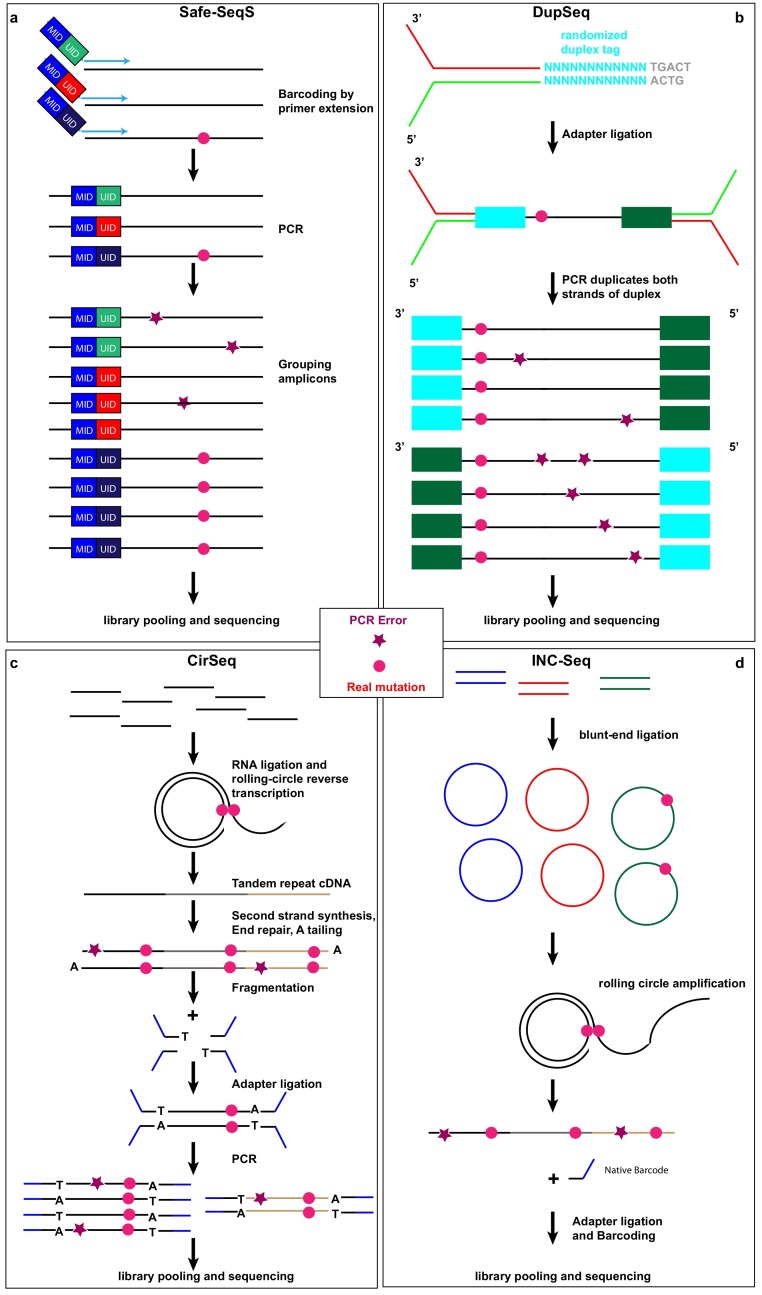
Fig. 1.
Library preparation approaches of consensus-based error correction for investigating virus quasispecies. (a) Safe-SeqS uses primers linked to unique molecular identifiers (UIDs) and mouse identifiers (MIDs) for reverse transcription, which not only enables the recognition of every original viral RNA strand after PCR amplification, but also allows multiplexing of samples in the same sequencing run. (b) DupSeq applies randomized duplex tags to each double-stranded DNA molecule in a way that derivative PCR products of the two strands can be informatively related to each other but also distinguishable. Consensus wild-type or mutation sequences are reached only if the reads of each of the double strands show identical sequences. (c) CirSeq begins by circularizing of single-stranded DNA fragments without any exogenous molecular barcodes followed by rolling-circle amplification, fragmentation and sequencing. (d) INC-Seq also entails circularization single-stranded DNA fragments followed by rolling-circle amplification of the loop; however, the end product is a long DNA strand (>10Kb) comprising concatenated copies of one of the strands of the starting molecule to be sequenced on a long-read platform. For INC-Seq, only in-silico fragmentation is performed for analysis following sequencing. For CirSeq and INC-seq, the random fragmentation points of the starting molecules serve as endogenous UIDs for consensus-based error correction. For all above-mentioned four methods, after library preparation, pooling and sequencing, sequences originating from the same viral RNA strand of the same sample, are collapsed to a single consensus sequence. True mutations (pink circle) can be distinguished from PCR errors (purple star). Due to limited space, sequencing errors are not marked here.