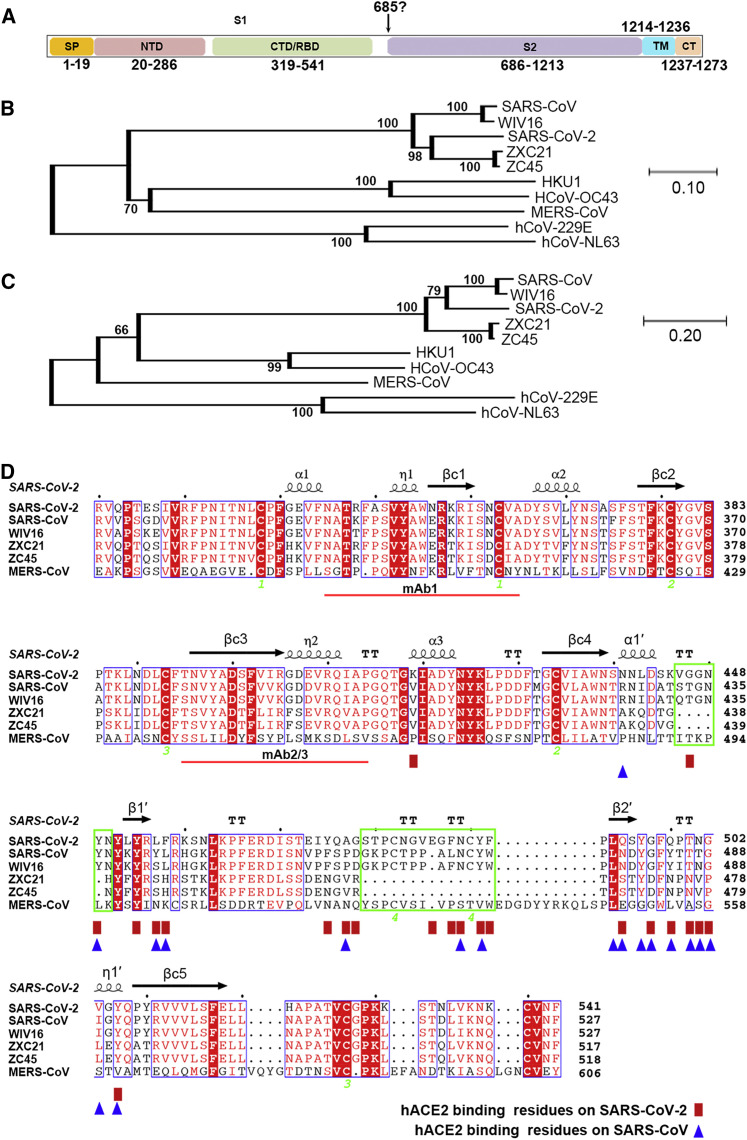
Figure S1.
Phylogenetic Analysis of SARS-CoV-2 and Sequence Alignments at the CTD Region, Related to Figures 2 and 3
(A) Schematic representation of the SARS-CoV-2 S protein based on the SARS-CoV S protein.
(B) Phylogenetic tree generated using MEGA (Tamura et al., 2013) with the S protein sequences.
(C) Phylogenetic tree generated using MEGA (Tamura et al., 2013) with the CTD region.
(D) Structure-based sequence alignment. The secondary structure elements were defined based on an ESPript (Robert and Gouet, 2014) algorithm and are labeled based on the SARS-CoV-2-CTD structure reported in this study. Spiral lines indicate α or 310 helices, and arrows represent β strands. The Arabic numerals 1-4 indicate cysteine residues that pair to form disulfide bonds. The red rectangles and blue triangles indicate the residues in the SARS-CoV-2-CTD and the SARS-RBD that interact with hACE2, respectively. Two deletions present in the ZXC21 and ZC45 external subdomains were highlighted with green boxes. The red lines indicate the epitopes recognized by mAb1 or mAb2/3.