Figure 5.
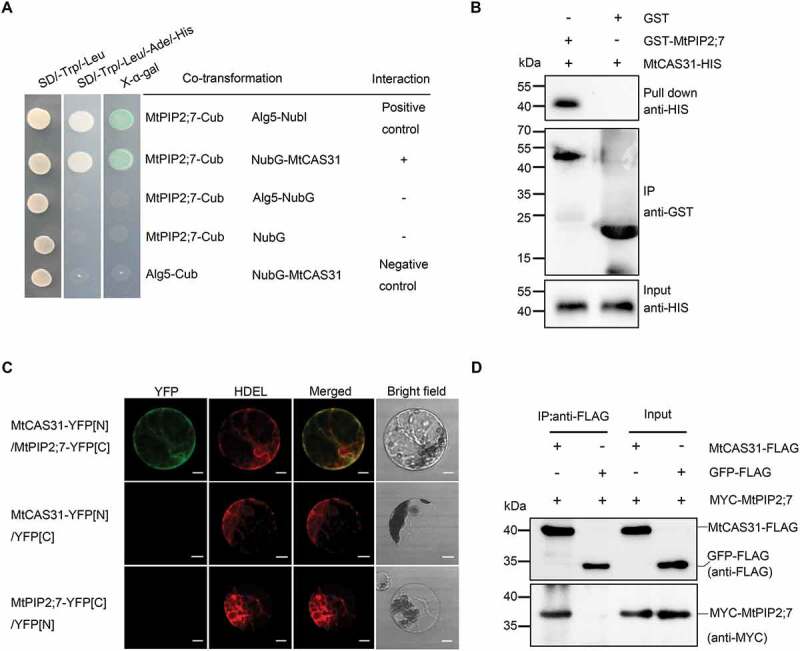
MtCAS31 interacts with MtPIP2;7. (A) Interaction of MtCAS31 and MtPIP2;7 in the transformed yeast strain NMY51, determined by a split-ubiquitin yeast two-hybrid assay. MtPIP2;7 was fused with the C terminus of ubiquitin (MtPIP2;7-Cub), and MtCAS31 was fused with the N terminus of mutant ubiquitin (NubG-MtCAS31). MtPIP2;7-Cub/Alg5-NubI was used as the positive control. MtPIP2;7-Cub/Alg5-NubG, MtPIP2;7-Cub/NubG and Alg5-CubI/NubG-MtCAS31 were employed as negative controls. Different co-transformed yeast cells were dropped onto synthetic dropout (SD) medium lacking tryptophan, leucine, adenine, and histidine (SD/-Trp/-Leu/-Ade/-His) and containing 20 mg/mL X-α-gal. (B) GST affinity-isolation assay to identify the interaction between MtPIP2;7 and MtCAS31. MtPIP2;7 was fused with a GST tag (GST-MtPIP2;7), and MtCAS31 was fused with a His tag (MtCAS31-His). GST-MtPIP2;7 or GST alone was precipitated with glutathione Sepharose 4B agarose beads for 3 h and incubated with MtCAS31-His. The precipitates were separated via SDS-PAGE and analyzed by immunoblotting using anti-His and anti-GST antibodies. (C) Interaction of MtCAS31 with MtPIP2;7 as determined by bimolecular fluorescence complementation. MtCAS31 was fused with the N terminus of YFP (MtCAS31-YFP[N]), and MtPIP2;7 was fused with the C terminus of YFP (MtPIP2;7-YFP[C]). Both constructs were driven by CaMV35S. HDEL-RFP, an ER marker, was co-transformed into Arabidopsis protoplasts. Co-transformed protoplasts were incubated for 16 h, and the fluorescence signals were detected by confocal laser scanning microscopy (Olympus FluoView FV1000) with excitation at 488 nm (for GFP fluorescence detection) and 546 nm (for RFP detection). MtCAS31-YFP[N]/Medtr4g415300-YFP[C]/HDEL-RFP and MtCAS31-YFP[N]/Medtr1g095070-YFP[C]/HDEL-RFP were employed as negative controls. YFP, yellow fluorescent protein. HDEL, amino acid sequence for localization to the ER. RFP, red fluorescent protein. Bar: 10 μm. (D) Detection of the MtCAS31-MtPIP2;7 interaction by coimmunoprecipitation (Co-IP). MtPIP2;7 was tagged with MYC (MtPIP2;7-MYC). GFP-FLAG/MtPIP2;7-MYC was employed as the negative control. Total proteins were extracted from the N. benthamiana leaf, which was co-transformed with the indicated constructs and incubated with FLAG beads to immunoprecipitate the target protein. Coprecipitated proteins were analyzed by immunoblotting using anti-FLAG and anti-MYC antibodies.
