Abstract
DNA ligase I and DNA ligase III/XRCC1 complex catalyze the ultimate ligation step following DNA polymerase (pol) β nucleotide insertion during base excision repair (BER). Pol β Asn279 and Arg283 are the critical active site residues for the differentiation of an incoming nucleotide and a template base and the N-terminal domain of DNA ligase I mediates its interaction with pol β. Here, we show inefficient ligation of pol β insertion products with mismatched or damaged nucleotides, with the exception of a Watson–Crick-like dGTP insertion opposite T, using BER DNA ligases in vitro. Moreover, pol β N279A and R283A mutants deter the ligation of the promutagenic repair intermediates and the presence of N-terminal domain of DNA ligase I in a coupled reaction governs the channeling of the pol β insertion products. Our results demonstrate that the BER DNA ligases are compromised by subtle changes in all 12 possible noncanonical base pairs at the 3′-end of the nicked repair intermediate. These findings contribute to understanding of how the identity of the mismatch affects the substrate channeling of the repair pathway and the mechanism underlying the coordination between pol β and DNA ligase at the final ligation step to maintain the BER efficiency.
INTRODUCTION
The base excision repair (BER) pathway is the predominant DNA repair mechanism in the cellular defense against small single-base DNA lesions such as DNA alkylation and oxidation products and involves many enzymatic steps proceeding in an orderly manner via protein-protein interactions (1–3). The repair pathway is initiated by the DNA damage-specific DNA glycosylase-mediated removal of a single-base lesion via hydrolysis of the N-glycosidic bond, which results in an abasic or apurinic/apyrimidinic (AP) site in double-stranded DNA (4). AP endonuclease 1 (APE1) then cleaves the phosphodiester backbone at the AP site, and leaves 3′-hydroxyl (3′-OH) and 5′-deoxyribosephosphate (5′-dRP) groups (5). DNA polymerase (pol) β exhibits two critical enzymatic activities during the BER pathway: removal of a 5′-dRP group by its 8-kDa lyase domain and filling of a template-directed gap by its 31-kDa polymerase domain (6–8). These pol β products generate a substrate for DNA ligation, which is the final step of the BER pathway and involves the sealing of the nicked DNA intermediate by DNA ligase I or DNA ligase III/X-ray repair cross-complementing protein 1 (XRCC1) complex (9,10). As indicated by biochemical and structural studies, the BER pathway functions via a substrate channeling mechanism that includes coordination of the substrate-product handoff between repair proteins (11–13). This mechanism prevents the release and accumulation of toxic and mutagenic single-strand break intermediates that could trigger cell-cycle arrest, apoptosis and harmful nuclease or recombination activities (14–16). However, how BER enzymes function together in a multiprotein/DNA complex to facilitate the channeling of toxic DNA repair intermediates remains unclear (17). Our previous studies revealed that pol β and BER DNA ligases coordinate with each other during the processing of the oxidative DNA damage product 7,8-dihydro-8′-oxo-dGTP (8-oxodGTP) (18–20).
Kinetic, structural, and computational studies have shown that pol β binds to one-nucleotide gapped DNA in an open conformation to form a binary complex (21–23). Upon subsequent nucleotide binding, the enzyme undergoes a conformational change to form the precatalytic closed ternary complex with two active-site catalytic and nucleotide-binding Mg2+ ions (24,25). In the presence of a Watson-Crick base pairing, the complex is optimized for DNA synthesis, which is known as an induced-fit mechanism where the binding of the correct nucleotide with a matched primer terminus aligns the catalytic participants for optimal chemistry and distorts the mismatched terminus to prevent incorporation (26–29). For example, time-lapse snapshots of the catalytic intermediates have demonstrated that the pol β active site undergoes diverse mismatch-induced molecular adjustments that differ from the conformational changes that the enzyme normally undergoes in response to binding to correct nucleotide (30). The extent of these conformational distortions in the pol β active site is dependent on the architecture of the mismatched template primer (31–34). However, how this architecture in a multiprotein/DNA complex could affect the BER process is poorly understood. In the present study, we evaluated the impacts of all possible mismatches and oxidative damage in DNA inserted by pol β on the substrate channeling of the coordinated repair pathway and fidelity of ligation catalyzed by BER DNA ligases (DNA ligase I and DNA ligase III/XRCC1 complex) in vitro.
Pol β is an error-prone polymerase that incorporates mismatches in approximately one of every 10,000 nucleotide insertion events during template-directed DNA synthesis in the BER pathway (21,25). Previous studies have shown that a high percentage of tumors have pol β variants with a mutation involving a single amino acid change in the active site of the enzyme, which results in an increased mismatch insertion rate and reduced fidelity (35,36). These cancer-associated pol β variants can lead to permanent cellular transformation, aberrant DNA repair and accumulation of toxic BER intermediates, and these findings suggest a pivotal role for pol β-mediated high-fidelity DNA synthesis in carcinogenesis (37,38). Therefore, the elucidation of the molecular mechanism underlying mismatch insertion coupled to ligation at the last step of the repair pathway is an important research topic that can be exploited to modulate repair and enhance human health.
DNA ligases catalyze the formation of a phosphodiester bond between adjacent 3′-OH and 5′-P termini of the repair intermediate and require adenosine triphosphate (ATP) and a divalent metal ion (Mg2+) for catalysis (39,40). Successful ligation relies on the formation of a Watson–Crick base pair between the 5′- and 3′-ends of the nicked DNA (41,42). The formation of a correct nucleotide insertion product that shows a stable closed pol β ternary complex with a matched template primer could enable recognition of the insertion product and efficient hand off of the resulting nicked repair intermediate from pol β to DNA ligase in the BER pathway. However, mismatched template-primer distortions at the repair intermediate could impact the ligation efficiency at the last step of the repair pathway. The multidomain architecture of DNA ligases allows these enzymes to sense the 3′ margin of the nick, and in the presence of 3′-damaged or modified DNA ends, DNA ligases might fail, resulting in the formation of 5′-adenylated DNA (43). These DNA intermediates with the 5′-AMP block could become persistent DNA strand breaks and mutagenic repair intermediates, which would lead to collapsed replication forks if not repaired by DNA end-processing enzymes (44,45).
In the present study, we investigated the ligation of pol β insertion products with matched, mismatched and 8-oxodG-containing primer termini by BER DNA ligases (DNA ligase I and DNA ligase III/XRCC1 complex) in a coupled reaction including pol β and DNA ligase in vitro. We found that the pol β-mismatched substrate complex could not be recognized by DNA ligase, whereas the pol β Watson-Crick-like dGTP:T insertion product was efficiently ligated similar to the nicked repair intermediate following pol β-matched dATP:T insertion. In contrast, pol β 8-oxodGTP insertion resulted in ligation failure. Moreover, the mutations in the side chains of the polymerase active site (N279A and R283K or R283A) completely abolished the ligation in a coupled reaction. We also showed that the N-terminal domain of DNA ligase I, which is responsible for the interaction with pol β, facilitates the channeling of pol β insertion products. Our results demonstrated that BER DNA ligases exhibited a distinct ability to ligate nicked repair intermediates with 3′-preinserted mismatches depending on the chemical characteristics of all 12 possible noncanonical base pairs between a primer terminus and a template base.
MATERIALS AND METHODS
Materials
Oligodeoxyribonucleotides with and without a 6-carboxyfluorescein (FAM) label were obtained from Integrated DNA Technologies. Solutions of the deoxynucleotides (dNTPs) dATP, dCTP, dGTP and dTTP were obtained from New England Biolabs.
Protein purifications
Human DNA polymerase β and DNA ligase I were purified as previously described (19,46,47). Briefly, the recombinant wild-type pol β and DNA ligase I proteins were overexpressed in One Shot BL21(DE3)pLysS Escherichia coli cells (Invitrogen) and grown at 37°C, and expression was induced once the cells reached an OD of 0.6. The cells were then grown overnight at 16°C. After centrifugation, the cells were lysed at 4°C by sonication in lysis buffer containing 25 mM HEPES (pH 7.5), 500 mM NaCl, 0.1% NP40 and a protease inhibitor cocktail. The lysate was pelleted at 10 444 × g for 1 h and then clarified by filtration. The pol β supernatant was loaded onto a GSTrap HP column (GE Health Sciences) and purified with elution buffer containing 50 mM Tris–HCl (pH 8.0) and 10 mM reduced glutathione. The collected fractions containing pol β were subsequently passed through a HiTrap Desalting HP column in a buffer containing 150 mM NaCl and 20 mM NaH2PO4 (pH 7.0). The DNA ligase I supernatant was loaded onto a HisTrap HP column (GE Health Sciences) and purified by elution with an increasing imidazole gradient (0–500 mM) at 4°C. The collected fractions containing DNA ligase I were subsequently loaded onto a HiTrap Heparin HP column (GE Health Sciences) with a linear gradient of NaCl up to 1 M. The resulting pure fractions of pol β and DNA ligase I were combined, concentrated with a centrifugal filter unit and stored in aliquots at −80°C.
The wild-type C- (261–918 aa) and N-terminal (1–259 aa) domains of human DNA ligase I and the full-length human DNA ligase I mutant (E566K) were cloned into the pET-24b expression vector (Novagen) using the primers listed in Supplementary Table S1. His-tagged recombinant proteins were overexpressed and purified as described above. Briefly, the cells were overexpressed in One Shot BL21(DE3) E. coli cells (Invitrogen) and grown at 37°C in TB medium, and expression was induced with 1.0 mM isopropyl β-D-thiogalactoside (IPTG) once the cells reached an OD of 1.0. The cells were grown overnight at 28°C, harvested and lysed at 4°C by sonication in lysis buffer containing 50 mM Tris–HCl (pH 7.0), 500 mM NaCl, glycerol 5% and 20 mM imidazole. The lysate was pelleted at 16 000 rpm for 40 min and then clarified by filtration. The supernatant was loaded onto a HisTrap HP column (GE Health Sciences) and purified by elution with an increasing imidazole gradient (0–500 mM) at 4°C. The collected fractions were subsequently loaded onto a Superdex200 gel-filtration column (GE Health Sciences) with buffer containing 50 mM Tris–HCl (pH 7.0), 500 mM NaCl and glycerol 5%. The collected fractions were loaded onto a Resource Q column (GE Health Sciences) with a linear gradient of NaCl up to 1 M. For all proteins, the resulting pure fractions were combined, concentrated with a centrifugal filter unit and stored in aliquots at −80°C.
Preparation of DNA substrates
The 1-nt-gapped and nicked DNA substrates were prepared as described previously (19). Briefly, the upstream primer (17-mer) with an FAM label at the 5′-end and the downstream primer (16-mer) were annealed in the presence of the template oligonucleotide (34-mer) to prepare the one-nucleotide-gapped DNA substrates for the nucleotide insertion assay. The upstream primer (17-mer) with an FAM label at the 5′-end and the downstream primer (16-mer) with an FAM label at the 3′-end were annealed in the presence of the template oligonucleotide (34-mer) to prepare the 1-nt-gapped DNA substrates for the coupled assays. The upstream primer (18-mer) and the downstream primer (16-mer) with an FAM label at the 3′-end were annealed in the presence of the template oligonucleotide (34-mer) to prepare the nicked DNA substrates for DNA ligation assays. The sequence information for all the double-stranded DNA substrates used in this study is presented in Supplementary Tables S2–4.
Nucleotide insertion assay
The nucleotide insertion assays were performed under steady-state conditions as described previously (19). One-nucleotide-gapped DNA substrates with template bases A, T, G and C were used (Supplementary Table S2). Briefly, the reaction mixture contained 50 mM Tris–HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT 100 μg ml−1 bovine serum albumin (BSA), 10% glycerol, DNA substrate (500 nM) and dGTP, dCTP, dATP, or dTTP (100 μM) in a final volume of 10 μl. The reaction was initiated by the addition of pol β (10 nM) and was incubated at 37°C for the times indicated in the figure legends. The reaction products were mixed with an equal amount of gel loading buffer (95% formamide, 20 mM ethylenediaminetetraacetic acid, 0.02% bromophenol blue and 0.02% xylene cyanol) and then separated by electrophoresis on an 18% polyacrylamide gel as described previously (19). The gels were scanned with a Typhoon PhosphorImager (Amersham Typhoon RGB), and the data were analyzed using ImageQuant software.
Nucleotide insertion coupled to ligation assay
The coupled assays were performed under steady-state conditions as described previously (19). One-nucleotide-gapped DNA substrates with template bases A, T, G and C were used (Supplementary Table S3). Briefly, the reaction was initiated by the addition of pol β (10 nM) and DNA ligase I or DNA ligase III/XRCC1 complex (10 nM) to a mixture (final volume of 10 μl) that contained 50 mM Tris–HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 100 μg ml−1 BSA, 10% glycerol, the DNA substrate (500 nM) and dGTP, dCTP, dATP, or dTTP (100 μM). The reaction mixtures were then incubated at 37°C for the times indicated in the figure legends and were quenched by mixing with an equal volume of loading dye. The products were separated, and the data were analyzed as described above.
DNA ligation assay
The ligation assays were performed under steady-state conditions as described previously (19), and nicked DNA substrates with preinserted 3′-dA, dC, dG or dA opposite template bases A, T, G and C were used (Supplementary Table S4). Briefly, the reaction was initiated by the addition of DNA ligase I or the DNA ligase III/XRCC1 complex (10 nM) to a mixture containing 50 mM Tris–HCl (pH 7.5), 100 mM KCl, 10 mM MgCl2, 1 mM ATP, 1 mM DTT, 100 μg ml−1 BSA, 10% glycerol and the DNA substrate (500 nM) in a final volume of 10 μl. The reaction mixture was incubated at 37°C and stopped at the time points indicated in the figure legends by mixing with an equal volume of loading dye. The reaction products were analyzed as described above.
RESULTS AND DISCUSSION
Impact of pol β template-primer mismatches on the ligation of repair intermediates
BER involves the channeling of repair intermediates from a nucleotide insertion by pol β to ligation of the 5′- and 3′-DNA ends of the resulting nicked product by DNA ligase I or DNA ligase III/XRCC1 complex (17,18). To understand this handoff from pol β with the matched versus mismatched template primer to DNA ligase, we assessed the rate of nucleotide insertion coupled to DNA ligation in vitro in a reaction mixture including pol β, DNA ligase and dNTP (dATP, dTTP, dGTP or dCTP) (Figure 1B). For this purpose, we used 1-nt gapped DNA substrates with template A, T, G or C (Supplementary Tables S2 and 3).
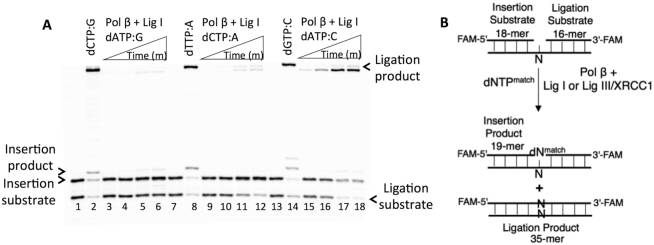
Figure 1.
Impact of pol β mismatch insertions on the ligation of repair intermediates by DNA ligase I. (A) Lanes 1, 7 and 13 are the negative enzyme controls of the one-nucleotide gapped DNA substrates Gcoupled, Acoupled and Ccoupled, respectively. Lanes 2, 8 and 14 show the positive controls for the insertion coupled to ligation products for dCTP:G, dTTP:A and dGTP:C, respectively. Lanes 3–6, 9–12 and 15–18 show the insertion coupled to ligation products for dATP:G, dCTP:A and dATP:C mismatches, respectively, obtained at the time points 0.5, 1, 3 and 5 min. (B) Illustrations of the 1-nt gapped DNA substrate with template A, T, G or C (N) and the reaction products from insertion and ligation observed in the coupled reaction including a matched dNTP.
We first evaluated the ligation of the pol β mismatch insertion products by DNA ligase I in a coupled reaction (Figure 1). The results revealed complete ligation of the pol β dCTP:G correct insertion (Figure 1A, lane 2) but no ligation of the pol β dATP:G insertion product (Figure 1A, lanes 3–6). With dCTP:A, we obtained weak pol β insertion and negligible accumulation of the ligation products (Figure 1A, lanes 8 versus 9–12). Interestingly, in both cases, the products of mismatch insertion coupled to ligation were mainly the ligation products of the one-nucleotide gapped DNA itself, as revealed by the difference in the size of the products (Supplementary Figure S1A and B, respectively). In contrast, for dATP:C mismatch insertion, DNA ligase I was able to very efficiently ligate the 1-nt gap with template C itself (Figure 1A, lanes 15–18), whereas complete ligation products were obtained with pol β dGTP:C insertion (Figure 1A, lane 14). These self-ligation products were observed with all pol β mismatches opposite C (Supplementary Figure S1C). The coupled reaction including pol β and DNA ligase III/XRCC1 complex yielded similar results for the above-mentioned template G, A and C mismatches, and a relatively weak amount of ligation products was obtained in all cases (Supplementary Figure S2).
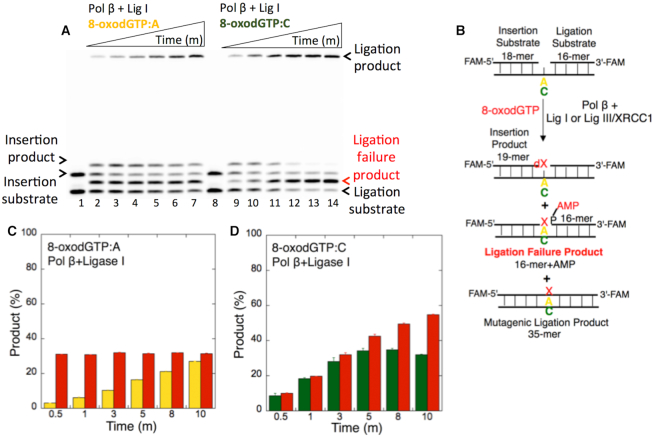
We subsequently tested pol β 8-oxodGTP insertion coupled to ligation using 1-nt gapped DNA substrates with template A or C (Figure 2B) as reported (19). In contrast to the findings obtained with undamaged DNA base mismatches (Figure 1), this experiment resulted in ligation failure, as revealed by the formation of the 5′-adenylate product, i.e. addition of AMP to the 5′-end of the substrate (Figure 2A). This ligation failure was accompanied by mutagenic ligation products for both pol β insertions, namely, 8-oxodGTP opposite A (Figure 2A, lanes 2–7) and 8-oxodGTP opposite C (Figure 2A, lanes 9–14), over the incubation time (Figure 2C and 2D).
Figure 2.
Ligation failure of pol β 8-oxodGTP insertion products. (A) Lanes 1 and 8 are the negative enzyme controls of the 1-nt gapped DNA substrates Acoupled and Ccoupled, respectively. Lanes 2–7 and 9–14 show the insertion coupled to ligation products from 8-oxodGTP:A and 8-oxodGTP:C, respectively, obtained at the time points 0.5, 1, 3, 5, 8 and 10 min. (B) Illustrations of the 1-nt gapped DNA substrate with template A or C and the insertion, ligation and ligation failure reaction products obtained in the coupled reaction including 8-oxodGTP. (C andD) The graphs show the time-dependent changes in the ligation (yellow for 8-oxodGTP:A and green for 8-oxodGTP:C) and ligation failure (red) products. The data are presented as the averages from three independent experiments ± SDs.
Our findings highlight the effect of the differences in the architecture of the pol β-mismatch primer template on the channeling of the resulting repair product to the subsequent DNA ligation step in the BER pathway. We showed that the product obtained from pol β insertion of mismatches opposite templates G, A and C cannot be used as a substrate for nick sealing by DNA ligase. This step could thus serve as a proofreading checkpoint for efficient substrate channeling in the BER pathway because it provides an opportunity for a proofreading enzyme, APE1, to excise the misinserted base (5). In contrast, DNA ligase attempts to ligate the repair intermediate with 8-oxodG inserted by pol β, which results in ligation failure. In this case, DNA-end processing enzymes such as APTX could play a role in the cleaning of an AMP block at the 5′-end (19,44,45).
In the control reaction, we observed efficient ligation after the pol β-matched dTTP insertion opposite A by DNA ligase I and DNA ligase III/XRCC1 complex (Supplementary Figure S3). Our data suggest that the resulting nicked product obtained after pol β-matched nucleotide insertion facilitates its ligation during the subsequent DNA ligation step in the repair pathway.
Mutagenic ligation of pol β Watson–Crick-like dGTP:T insertion products
Incorrect dG:dT base pairing during DNA repair or replication, if left unrepaired, leads to transition or transversion point mutations and is believed to be a prominent source of base substitution errors in tumor suppressor genes in multiple forms of cancer (48,49). It has been shown that the pol β active site with a G-T mismatch escapes mismatch discrimination through ionization of the wobble base pair and exhibits a Watson–Crick-like conformation in a closed state (33,34). However, the mechanism through which these structural arrangements of pol β can affect the downstream steps of the BER pathway is unknown. To understand the channeling of the repair intermediate in a wobble vs Watson–Crick-like conformation from pol β to DNA ligase, we investigated dGTP insertion opposite T (i.e. dGTP:T) coupled to ligation in vitro (Figure 3B).
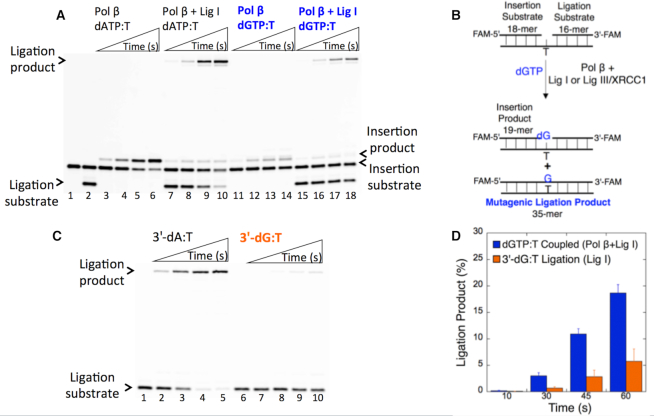
Figure 3.
Impact of Watson-Crick-like dGTP:T conformation on DNA ligation. (A) Ligation of pol β dGTP:T insertion products by DNA ligase I. Lanes 1 and 2 are the negative enzyme controls of the 1-nt gapped DNA substrates Tinsertion and Tcoupled, respectively. Lanes 3–6 and 11–14 show the pol β dATP:T and dGTP:T insertion products, respectively, obtained at the time points 10, 30, 45 and 60 s. Lanes 7–10 and 15–18 show the insertions coupled to ligation products obtained for dATP:T and dGTP:T, respectively, at the time points 10, 30, 45 and 60 s. (B) Illustrations of the 1-nt gapped DNA substrate with template T and the insertion and ligation reaction products observed in the coupled reaction including dGTP. (C) Ligation of the nicked DNA with preinserted 3′-dG:T. Lanes 1 and 6 are the negative enzyme controls of the nicked DNA substrate Tligation with 3′-dA and 3′-dG, respectively. Lanes 2–5 and 7–10 show the ligation reaction products obtained for 3′-dA:T and 3′-dG:T, respectively, at the time points 10, 30, 45 and 60 s. (D) The graph shows the time-dependent changes in the ligation products obtained with a coupled (blue, dGTP:T) versus ligation (orange, 3′-dG:T) reaction. The data are presented as the averages from three independent experiments ± SDs.
In a coupled reaction including pol β and DNA ligase I, the products of dGTP:T insertion (Figure 3A, lanes 11–14) and the corresponding complete ligation (Figure 3A, lanes 15–18) were obtained at earlier time points (10–60 s) compared with those obtained with all other mismatches (0.5–5 min) tested in this study (Figure 1). These products are similar to those obtained by pol β-matched dATP:T insertion (Figure 3A, lanes 3–6) and insertion coupled to complete ligation (Figure 3A, lanes 7–10). Similarly, in a coupled reaction including the DNA ligase III/XRCC1 complex, we observed complete ligation of the pol β dGTP:T insertion products (Supplementary Figure S4A) but the product amount was relatively lower than that obtained with DNA ligase I (Supplementary Figure S4B).
The ligation of the nicked repair intermediate by DNA ligase alone should be considered when interpreting these results. DNA ligase I itself was unable to ligate the nicked DNA with a 3′-preinserted dG:T mismatch (Figure 3C, lanes 7–10), whereas the efficient ligation of 3′-preinserted dA:T (Figure 3C, lanes 2–5) was observed at the same time points (10–60 s). The quantification of the ligation products from the pol β insertion coupled to ligation (dGTP:T) vs DNA ligase itself (3′-dG:T) revealed that the pol β Watson–Crick-like dG:T conformation enables ligation of the resulting nicked repair intermediate (Figure 3D). Similarly, in a ligation reaction including the DNA ligase III/XRCC1 complex, we observed fewer ligation products of the nicked substrate with 3′-dG:T (Supplementary Figure S4C).
Our findings demonstrated that the BER DNA ligases efficiently ligate the pol β insertion products in the dG:T conformation that fit well within the dimensions of the double helix to preserve the geometry of Watson–Crick base pairs in a manner similar to that observed with the complete ligation of the stable pol β closed-product complex with dA:T base pair. These ligation products could be mutagenic repair intermediates. Moreover, this inaccuracy (i.e. dG-T mismatch insertion coupled to efficient ligation) at the end of the repair pathway could be a fundamental source of genomic instability that has deleterious biological consequences.
Effect of pol β active-site mutants on the ligation of dGTP:T mismatch products
It has been reported that the formation of Watson-Crick-like dG:T base pairs in the closed conformation state at the pol β active site is induced by Arg283 and Asn279 residues that mediate the interactions in the DNA minor groove with a template base and an incoming nucleotide, respectively (50–53). Based on the above-described results, we subsequently examined the mutations in Arg283 and Asn279 to understand their effects on the ligation of pol β dG:T insertion products.
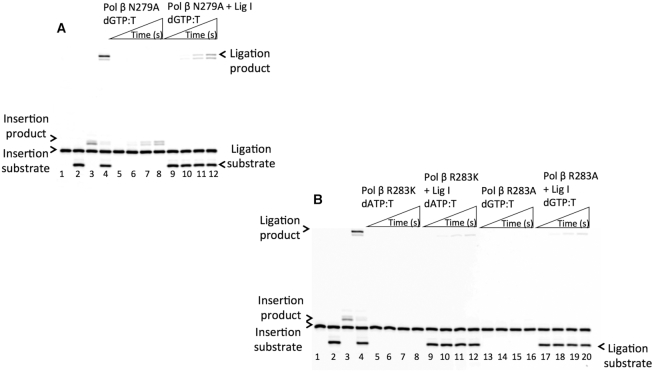
The levels of dGTP:T insertion (Figure 4A, lanes 5–8) and the products of insertion coupled to ligation (Figure 4A, lanes 9–12) obtained with pol β N279A were lower than those with wild-type pol β (Figure 4A, lanes 3 and 4). We observed complete disruption in the levels of dGTP:T insertion (Figure 4B, lanes 5–8 and 13–16) and the products of insertion coupled to ligation (Figure 4B, lanes 9–12 and 17–20) with R283K and R283A, respectively, at the same time points (10–60 s) compared with the results with wild-type pol β (Figure 4B, lanes 3 and 4). Similarly, we observed a lack of channeling of the repair intermediate in the presence of the same pol β active-site mutants inserting dGTP:T in a coupled reaction including the DNA ligase III/XRCC1 complex (Supplementary Figure S5). Furthermore, we investigated the impact of the Asp256 side chain using the pol β D256A mutant. These results served as a negative control, which indicated that the D256A enzyme could not stabilize the incoming dGTP opposite T (Supplementary Figure S6, lanes 13–20), and this outcome was not surprising because Asp256 is required for binding of the active-site catalytic metal that is essential for the nucleotidyl transfer reaction (27), as shown by a lack of enzyme activity for the correct dATP:T base pair in comparison to wild-type pol β (Supplementary Figure S6, lanes 3–4 versus 5–12). These results confirmed the importance of hydrogen bonding to the minor groove edge of an incoming nucleotide and the transient nature of spontaneous dG:T replication errors. Our findings suggest that DNA ligase can sense and respond to the distinct environment that the staggered pol β conformation creates due to mutations in the enzyme active site, which discourages the ligase from joining the DNA ends of the repair intermediate.
Figure 4.
Effect of pol β active-site mutations on the ligation of dGTP:T insertion products by DNA ligase I. In both panels, lanes 1 and 2 are the negative enzyme controls of the one-nucleotide gapped DNA substrates Tinsertion and Tcoupled, respectively. Lanes 3 and 4 show the positive controls for the wild-type pol β dGTP:T insertion and insertion coupled to ligation products, respectively. (A) Lanes 5–8 and 9–12 show the pol β N279A dGTP:T insertion and insertion coupled to ligation products, respectively, obtained at the time points 10, 30, 45 and 60 s. (B) Lanes 5–8/9–12 and 13–16/17–20 show the pol β R283K and R283A dGTP:T insertion/insertion coupled to ligation products, respectively, obtained at the time points 10, 30, 45 and 60 s. The gels are representative of three independent experiments.
Impact of DNA ligase I interaction on the ligation of pol β insertion products
The molecular machinery responsible for the BER pathway relies on coordination of the repair enzymes with each other to receive the damaged DNA substrate and efficiently pass the resulting repair product through a highly coordinated process to repair simple base lesions (11–16). The N-terminal region of DNA ligase I has no catalytic residues and participates in protein-protein interactions (17). Thermodynamic, domain mapping and coimmunoprecipitation studies have shown that this part of the ligase mediates its interaction with pol β (54,55).
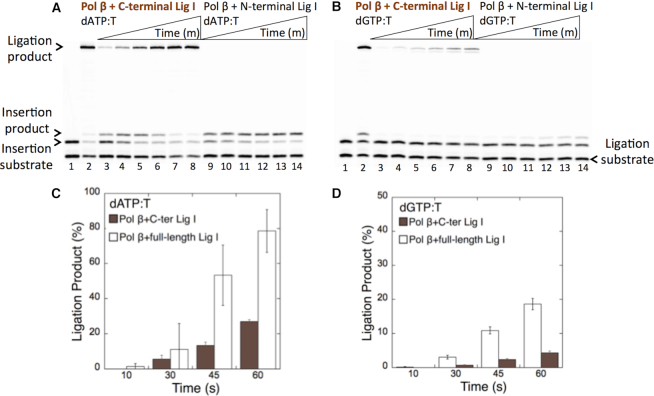
The above-described results indicate that the pol β active site can accommodate the dG:T base pair, and the pol β-dGTP:T ternary complex is able to serve as a substrate in a manner similar to that of the pol β-dATP:T complex, which enables both DNA synthesis and ligase activities. To further analyze the effect of this physical interaction on the ligation of pol β insertion products with the matched versus mismatched template primer, we performed coupled reactions using the N- and C-terminal domains of DNA ligase I and full-length pol β.
As expected, we did not observe any ligation products in a coupled reaction including the N-terminal domain of DNA ligase I and pol β for both correct dATP:T (Figure 5A, lanes 9–14) and mismatch dGTP:T (Figure 5B, lanes 9–14) insertions. In a coupled reaction including the C-terminal domain of DNA ligase I and pol β, the ligation products of dATP:T (Figure 5A, lanes 3–8) and dGTP:T (Figure 5B, lanes 3–8) insertions were obtained. In contrast, the amount of ligation products obtained in a coupled reaction including the C-terminal domain of the ligase was ∼4-fold lower compared with that obtained with full-length DNA ligase I (Figures 5C and 5D). This result suggests the requirement of pol β-DNA ligase I functional interaction for the efficient ligation of pol β insertion products and the channeling of the repair intermediates with inserted matched or mismatched bases in the BER pathway. Moreover, using the DNA ligase I mutant E566K, which is unable to form the ligase-AMP intermediate (56), we showed the requirement of catalytically active ligase in a coupled reaction for the channeling of pol β dATP:T insertion products (Supplementary Figure S7A). In the control ligation reactions with the nicked DNA substrate including correct 3′-preinserted dT:A, we confirmed that the DNA ligase I mutant E566K (Supplementary Figure S7B) showed a lack of ligation activity and found that the full-length and C-terminal domain of DNA ligase I exhibited the same ligase activity (Supplementary Figure S8).
Figure 5.
Effect of DNA ligase I interaction on the ligation of pol β dATP:T and dGTP:T insertion products. In both panels, lane 1 is the negative enzyme control of the one-nucleotide gapped DNA substrate Tcoupled, and lane 2 shows the positive control for the insertion coupled to the ligation product in the reaction including pol β and full-length DNA ligase I. (A) Lanes 3–8 and 9–14 show the pol β insertion coupled to ligation products obtained with the C- and N-terminal domains of DNA ligase I, respectively, in the presence of dATP:T at the time points 30, 45, 60, 120, 180 and 240 s. (B) Lanes 3–8 and 9–14 show the pol β insertion coupled to ligation products obtained with the C- and N-terminal domains of DNA ligase I, respectively, in the presence of dGTP:T at the time points 30, 45, 60, 120, 180 and 240 s. (C andD) The graph shows the time-dependent changes in the ligation products obtained in the coupled reactions including full-length versus the C-terminal domain of DNA ligase I for dATP:T (C) and dGTP:T (D). The data are presented as the averages from three independent experiments ± SDs.
Specificity of BER DNA ligases for the ligation of 3′-preinserted base mismatches
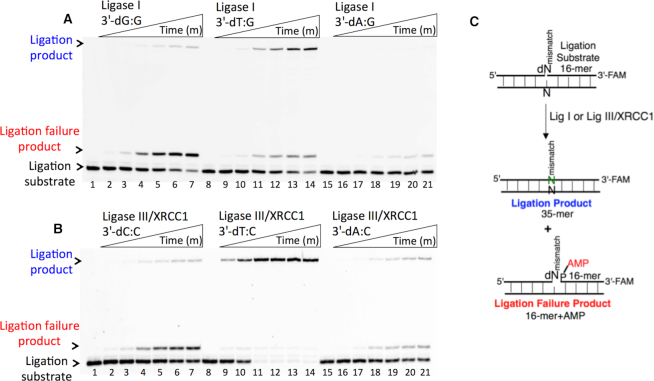
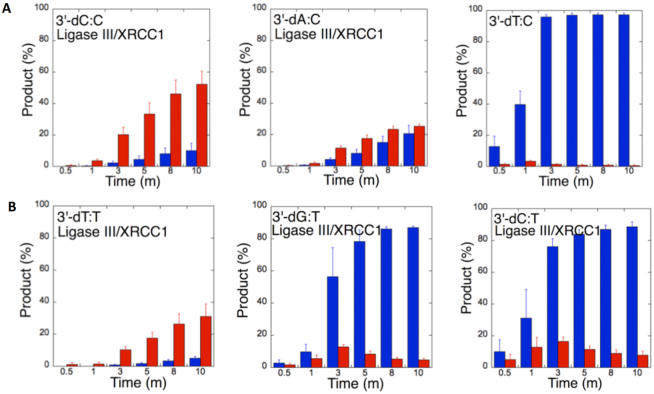
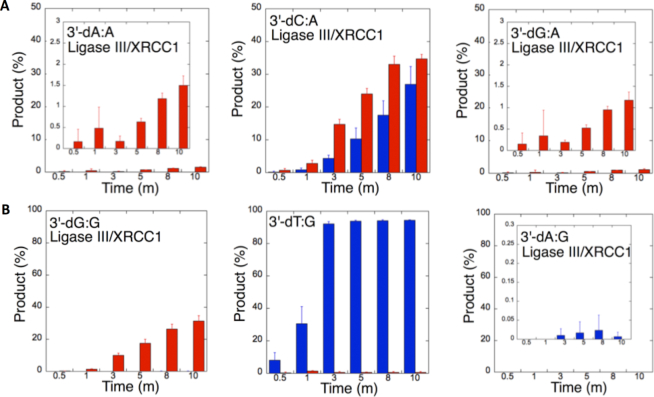
To understand the 3′-end structural surveillance of the BER DNA ligases (DNA ligase I and DNA ligase III/XRCC1 complex) for the ligation of the repair intermediates with 3′-preinserted mismatches that mimic the pol β mismatch insertion products described above, we assessed the ligation reaction in a mixture containing DNA ligase alone in vitro (Figure 6C). For this purpose, we used nicked DNA substrates with all 12 possible noncanonical base pairs, including 3′-preinserted dA, dT, dG, or dC opposite templates A, T, G or C (Supplementary Table S4).
Figure 6.
Ligation efficiency of the repair intermediates with 3′-preinserted mismatches by DNA ligase I and DNA ligase III/XRCC1 complex. (A) Lanes 1, 8 and 15 are the negative enzyme controls of the nicked DNA substrates with 3′-dG:G, 3′-dT:G and 3′-dA:G, respectively, and lanes 2–7, 9–14 and 16–21 show the reaction products obtained with DNA ligase I at the time points 0.5, 1, 3, 5, 8 and 10 min. (C) Illustrations of the nicked DNA substrate with template A, T, G or C (N) and the ligation and ligation failure reaction products obtained in the reaction including 3′-preinserted mismatches (dNmismatch). (B) Lanes 1, 8 and 15 are the negative enzyme controls of the nicked DNA substrates with 3′-dC:C, 3′-dT:C and 3′-dA:C, respectively, and lanes 2–7, 9–14 and 16–21 show the reaction products obtained with DNA ligase III/XRCC1 complex at the time points 0.5, 1, 3, 5, 8 and 10 min.
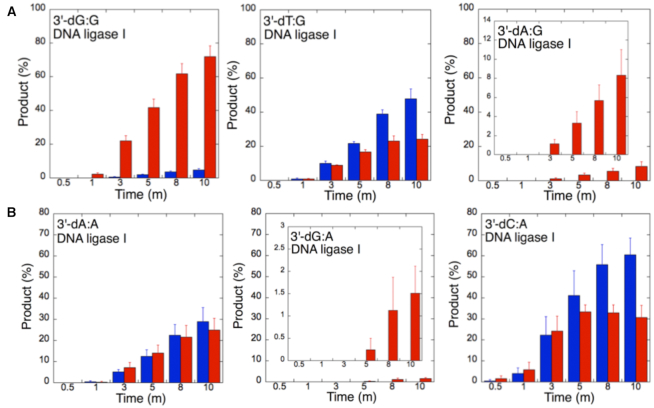
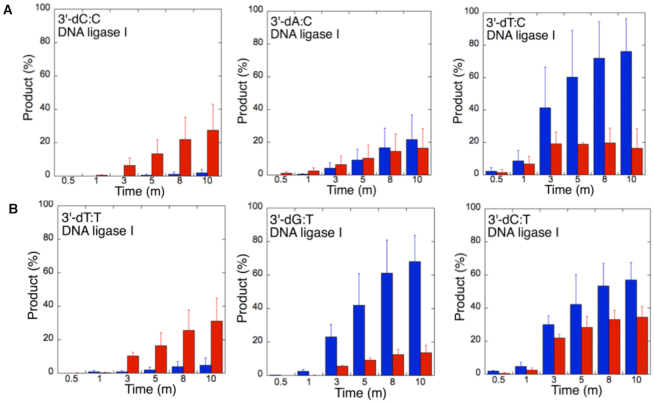
The ligation of the 3′-preinserted mismatches opposite G with DNA ligase I yielded high and low levels of ligation failure products for 3′-dG:G (Figure 6A, lanes 2–7) and 3′-dA:G (Figure 6A, lanes 16–21), respectively. We observed the products of both ligation and ligation failure for the nicked substrate with 3′-dT:G (Figure 6A, lanes 9–14) over the incubation time (Figure 7A). For the nicked substrate with 3′-preinserted mismatches opposite A (Figure 7B), the ligation of 3′-dA:A and 3′-dC:A resulted in the simultaneous appearance of ligation and ligation failure products at the same time points (Supplementary Figure S9A, lanes 2–7 and 16–21, respectively), but DNA ligase I was unable to ligate the nicked substrate with 3′-dG:A (Supplementary Figure S8A, lanes 9–14). The analysis of template C mismatches (Figure 8A) revealed ligation failure products for 3′-dC:C (Supplementary Figure S9B, lanes 2–7). However, both ligation and ligation failure products at different amounts were obtained for 3′-dT:C and 3′-dA:C (Supplementary Figure S9B, lanes 9–14 and 16–21, respectively). The ligation of 3′-preinserted mismatches opposite T (Figure 8B) mainly yielded ligation failure products for 3′-dT:T, (Supplementary Figure S10A, lanes 2–7), whereas ligation and ligation failure products appeared simultaneously for 3′-dG:T and 3′-dC:T (Supplementary Figure S10A, lanes 9–14 and 16–21, respectively).
Figure 7.
Mismatch specificity of DNA ligase I for the ligation of the repair intermediates with 3′-preinserted mismatches opposite templates G and A. The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Figure 6A and Supplementary Figure S9A.
Figure 8.
Mismatch specificity of DNA ligase I for the ligation of the repair intermediates with 3′-preinserted mismatches opposite templates C and T. The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Supplementary Figures S9B and 10A.
We also tested the specificity of the end-joining ability of the DNA ligase III/XRCC1 complex for all 12 non-canonical mismatches (Figures 9 and 10). The analysis of template C mismatches (Figure 9A) revealed complete ligation at earlier time points for 3′-dT:C (Figure 6B, lanes 9–14), whereas the other template C mismatches (3′-dC:C and 3′-dA:C) mainly produced ligation failure products (Figure 6B, lanes 2–7 and 16–21, respectively). For the nicked substrate with 3′-preinserted mismatches opposite T (Figure 9B), DNA ligase III/XRCC1 complex exhibited its highest activity with 3′-dG:T and 3′-dC:T (Supplementary Figure S10B, lanes 9–14 and 16–21, respectively). The ligation of 3′-preinserted mismatches opposite A (Figure 10A) showed that the DNA ligase III/XRCC1 complex exhibited no activity against almost all mismatches (Supplementary Figure S11A). The ligation of template G mismatches (Figure 10B) yielded significantly higher amounts of ligation products with 3′-dT:G (Supplementary Figure S11B, lanes 9–14), and we found no activity with 3′-dA:G (Supplementary Figure S11B, lanes 16–21). The control experiments confirmed complete ligation of the repair intermediates with 3′-preinserted Watson–Crick base pairings (3′-dT:A and 3′-dG:C) by DNA ligase I and the DNA ligase III/XRCC1 complex (Supplementary Figure S12).
Figure 9.
Mismatch specificity of DNA ligase III/XRCC1 complex for the ligation of the repair intermediates with 3′-preinserted mismatches opposite templates C and T. The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Figure 6B and Supplementary Figure S10B.
Figure 10.
Mismatch specificity of DNA ligase III/XRCC1 complex for the ligation of the repair intermediates with 3′-preinserted mismatches opposite templates A and G. The graphs show the time-dependent changes in the ligation (blue) and ligation failure (red) products, and the data are presented as the averages from three independent experiments ± SDs. The gel images are presented in Supplementary Figure S11.
Our results indicate that BER DNA ligases can ligate G:T mismatches with two hydrogen bonds and a base-pair size that is nearly indistinguishable from that of a Watson–Crick base pair, to a significant degree with high efficiency. The DNA ligase III/XRCC1 complex is able to join the ends with 3′-dG:T or 3′-dT:G and 3′-dT:C or 3′-dC:T much more efficiently than DNA ligase I (Figures 7–10 and Supplementary Figure S13). Interestingly, we also showed efficient ligation of pol β dGTP:T insertion products in a coupled reaction including both DNA ligase and pol β (Figure 3). In addition, our results indicate that the presence of a purine base on the repair intermediate, either in the template (3′-dA:G) or at the 3′-end (3′-dG:A), deters end joining when paired to another purine base by both BER DNA ligases (Figures 7 and 10). Overall, these results reveal that the BER DNA ligases recognize subtle base differences at either the template or the 3′-end when sealing nicked DNA, and the mismatch discrimination of the BER DNA ligases can vary depending on the DNA end structure of the repair intermediate (Supplementary Figure S13). Indeed, the substrate specificity of BER DNA ligases differs when the nicked repair intermediate with a non-canonical base pair is handed from the pol β-mediated mismatch insertion step. In this case, the interplay between pol β and DNA ligase in a multiprotein/DNA complex would be important for controlling the ligation of DNA repair intermediates at the last step of the BER pathway.
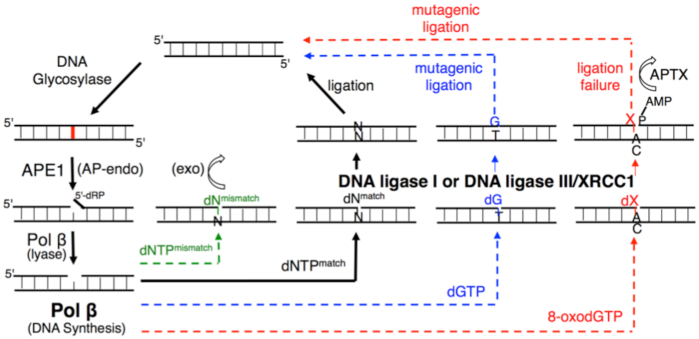
According to our model (Figure 11), we hypothesized that the pol β-substrate/product complex with a poor mismatch versus a good match geometry could serve as a structural fidelity checkpoint at which the channeling of the repair intermediate to the subsequent DNA ligation step in the BER pathway can be halted or facilitated. In the presence of a pol β matched-substrate/product that could protect the resulting nicked substrate, the DNA intermediate is efficiently ligated. Moreover, the architecture of a pol β ternary complex with a mismatch in the active site forms a premutagenic structural intermediate with mismatched termini that deters its ligation. This phenomenon could provide an opportunity for 3′-proofreading by APE1. However, the dG:T mismatch that adopts a Watson–Crick-like conformation in the pol β active site encourages mutagenic ligation. Finally, DNA ligase fails to catalyze the premutagenic DNA nick with oxidative DNA damage (8-oxodG) inserted by pol β, resulting in the formation of ligation failure or abortive repair intermediates, and these intermediates could give DNA-end processing enzymes (i.e. APTX) additional time to restore 5′-AMP ends.
Figure 11.
Illustration of the substrate channeling mechanism of the BER pathway in the presence of the pol β-substrate/product complex with matched, mismatched, and damaged DNA. The model shows the interplay between pol β and BER DNA ligases (DNA ligase I and DNA ligase III/XRCC1 complex) at the last step of the repair pathway, which serves as a structural fidelity checkpoint at which the channeling of the repair intermediate to the subsequent DNA ligation step could lead to efficient ligation, mutagenic ligation or ligation failure.
Although X-ray crystal structures of DNA ligases in complex with DNA have revealed similar ligation reactions through conserved three-domain core architectures, DNA ligases display different fidelity profiles depending on the type (human ligase I, III or IV) and source (human, Saccharomyces cerevisiae, T4 bacteriophage, archaea or virus) of DNA ligase (57–68). We also observed similar differences in the ligation efficiency of human BER DNA ligases depending on the identity of all 12 mismatches (Supplementary Figure S13). Therefore, structural studies of all mismatches tested in this study will be necessary to enhance our understanding of the mechanism underlying the mismatch discrimination of DNA ligases promoted by pol β-mediated mutagenesis and to elucidate the functional interaction between pol β and BER DNA ligases and its effect on the channeling of repair intermediates with various conformational architectures.
Supplementary Material
ACKNOWLEDGEMENTS
The author thanks Dr Alan E. Tomkinson (University of New Mexico) for providing the expression plasmid vector for human DNA ligase I and purified protein DNA ligase III/XRCC1 complex. The expression plasmid vector of wild-type DNA polymerase β and the purified mutant proteins were generous gifts from Samuel H. Wilson (NIEHS). The author thanks Dr Linda Bloom (University of Florida) for the helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institute of Environmental Health Sciences [R00ES026191]. Funding for open access charge: National Institute of Environmental Health Sciences [R00ES026191].
Conflict of interest statement. None declared.
REFERENCES
- 1. Krokan H.E., Nilsen H., Skorpen F., Otterlei M., Slupphaug G.. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000; 476:73–77. [DOI] [PubMed] [Google Scholar]
- 2. Lindahl T. Keynote: past, present, and future aspects of base excision repair. Prog. Nucleic Acid Res. Mol. Biol. 2001; 68:xvii–xxx. [DOI] [PubMed] [Google Scholar]
- 3. Parikh S.S., Mol C.D., Tainer J.A.. Base excision repair enzyme family portrait: integrating the structure and chemistry of an entire DNA repair pathway. Structure. 1997; 5:1543–1550. [DOI] [PubMed] [Google Scholar]
- 4. Srivastava D.K., Berg B.J., Prasad R., Molina J.T., Beard W.A., Tomkinson A.W., Wilson S.H.. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J. Biol. Chem. 1998; 273:21203–21209. [DOI] [PubMed] [Google Scholar]
- 5. Whitaker A.M., Flynn T.S., Freudenthal B.D.. Molecular snapshots of APE1 proofreading mismatches and removing DNA damage. Nat. Commun. 2018; 9:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beard W.A., Prasad R., Wilson S.H.. Activities and mechanism of DNA polymerase β. Methods Enzymol. 2006; 408:91–107. [DOI] [PubMed] [Google Scholar]
- 7. Beard W.A., Wilson S.H.. Structure and mechanism of DNA polymerase β. Biochemistry. 2014; 53:2768–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beard W.A., Wilson S.H.. Structure and mechanism of DNA polymerase β. Chem. Rev. 2006; 106:361–382. [DOI] [PubMed] [Google Scholar]
- 9. Tomkinson A.E., Vijayakumar S., Pascal J.M., Ellenberger T.. DNA ligases: structure, reaction mechanism, and function. Chem. Rev. 2006; 106:687–699. [DOI] [PubMed] [Google Scholar]
- 10. Sleeth K.M., Robson R.L., Dianov G.L.. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry. 2004; 43:12924–12930. [DOI] [PubMed] [Google Scholar]
- 11. Prasad R., Shock D.D., Beard W.A., Wilson S.H.. Substrate channeling in mammalian base excision repair pathways: passing the baton. J. Biol. Chem. 2010; 285:40479–40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prasad R., Williams J.G., Hou E.W., Wilson S.H.. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2012; 40:11571–11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Moor N.A., Vasil’eva I.A., Anarbaev R.O., Antson A.A., Lavrik O.I.. Quantitative characterization of protein-protein complexes involved in base excision DNA repair. Nucleic Acids Res. 2015; 43:6009–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson S.H., Kunkel T.A.. Passing the baton in base excision repair. Nat. Struct. Biol. 2000; 7:176–178. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y., Prasad R., Beard W.A., Kedar P.S., Hou E.W., Shock D.D., Wilson S.H.. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J. Biol. Chem. 2007; 282:13532–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Prasad R., Beard W.A., Batra V.K., Liu Y., Shock D.D., Wilson S.H.. A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways. Mol. Biol. 2011; 45:586–600. [PMC free article] [PubMed] [Google Scholar]
- 17. Çağlayan M. Interplay between DNA polymerases and DNA ligases: Influence on substrate channeling and the fidelity of DNA ligation. J. Mol. Biol. 2019; 431:2068–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Çağlayan M., Wilson S.H.. Oxidant and environmental toxicant-induced effects compromise DNA ligation during base excision DNA repair. DNA Repair (Amst.). 2015; 35:85–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Çağlayan M., Horton J.K., Dai D.P., Stefanick D.F., Wilson S.H.. Oxidized nucleotide insertion by pol β confounds ligation during base excision repair. Nat. Commun. 2017; 8:14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Çağlayan M., Wilson S.H.. Role of DNA polymerase β oxidized nucleotide insertion in DNA ligation failure. J. Radiat. Res. 2017; 58:603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beard W.A., Shock D.D., Vande Berg B.J., Wilson S.H.. Efficiency of correct nucleotide insertion governs DNA polymerase fidelity. J. Biol. Chem. 2002; 277:47393–47398. [DOI] [PubMed] [Google Scholar]
- 22. Beard W.A., Wilson S.H.. Structural insights into the origins of DNA polymerase fidelity. Structure. 2003; 11:489–496. [DOI] [PubMed] [Google Scholar]
- 23. Roettger M.P., Bakhtina M., Tsai M.D.. Mismatched and matched dNTP incorporation by DNA polymerase β proceed via analogous kinetic pathways. Biochemistry. 2008; 47:9718–9727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Batra V.K., Beard W.A., Shock D.D., Pedersen L.C., Wilson S.H.. Nucleotide-induced DNA polymerase active site motions accommodating a mutagenic DNA intermediate. Structure. 2005; 13:1225–1233. [DOI] [PubMed] [Google Scholar]
- 25. Beard W.A., Osheroff W.P., Prasad R., Sawaya M.R., Jaju M., Wood T.G., Kraut J., Kunkel T.A., Wilson S.H.. Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase β. J. Biol. Chem. 1996; 271:12141–12144. [DOI] [PubMed] [Google Scholar]
- 26. Johnson K.A. Role of induced fit in enzyme specificity: a molecular forward/reverse switch. J. Biol. Chem. 2008; 283:26297–26301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sawaya M.R., Prasad R., Wilson S.H., Kraut J., Pelletier H.. Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997; 36:11205–11215. [DOI] [PubMed] [Google Scholar]
- 28. Krahn J.M., Beard W.A., Wilson S.H.. Structural insights into DNA polymerase β deterrents for misincorporation support an induced-fit mechanism for fidelity. Structure. 2004; 12:1823–1832. [DOI] [PubMed] [Google Scholar]
- 29. Batra V.K., Beard W.A., Pedersen L.C., Wilson S.H.. Structures of DNA polymerase mispaired DNA termini transitioning to pre-catalytic complexes support an induced-fit fidelity mechanism. Structure. 2016; 24:1863–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Freudenthal B.D., Beard W.A., Shock D.D., Wilson S.H.. Observing a DNA polymerase choose right from wrong. Cell. 2013; 154:157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Batra V.K., Beard W.A., Shock D.D., Pedersen L.C., Wilson S.H.. Structures of DNA polymerase beta with active-site mismatches suggest a transient abasic site intermediate during misincorporation. Mol. Cell. 2008; 30:315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Beard W.A., Shock D.D., Yang X.P., DeLauder S.F., Wilson S.H.. Loss of DNA polymerase beta stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J. Biol. Chem. 2002; 277:8235–8242. [DOI] [PubMed] [Google Scholar]
- 33. Koag M.C., Nam K., Lee S.. The spontaneous replication error and the mismatch discrimination mechanisms of human DNA polymerase β. Nucleic Acids Res. 2014; 42:11233–11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahn J., Kraynov V.S., Zhong X., Werneburg B.G., Tsai M.D.. DNA polymerase β: effects of gapped DNA substrates on dNTP specificity, fidelity, processivity and conformational changes. Biochem. J. 1998; 331:79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shah A.M., Maitra M., Sweasy J.B.. Variants of DNA polymerase β extend mispaired DNA due to increased affinity for nucleotide substrate. Biochemistry. 2003; 42:10709–10717. [DOI] [PubMed] [Google Scholar]
- 36. Sweasy J.B., Lang T., Starcevic D., Sun K.W., Lai C.C., Dimaio D., Dalal S.. Expression of DNA polymerase β cancer-associated variants in mouse cells results in cellular transformation. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:14350–14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kelley M.R., Logsdon D., Fishel M.L.. Targeting DNA repair pathways for cancer treatment: what's new. Future Oncol. 2014; 10:1215–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallace S.S., Murphy D.L., Sweasy J.B.. Base excision repair and cancer. Cancer Lett. 2012; 327:73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ellenberger T., Tomkinson A.E.. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem. 2008; 77:313–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Timson D.J., Singleton M.R., Wigley D.B.. DNA ligases in the repair and replication of DNA. Mutat. Res. 2000; 460:301–318. [DOI] [PubMed] [Google Scholar]
- 41. Cherepanov A.V., Vries S.de. Dynamic mechanism of nick recognition by DNA ligase. Eur. J. Biochem. 2002; 269:5993–5999. [DOI] [PubMed] [Google Scholar]
- 42. Dickson K.S., Burns C.M., Richardson J.P.. Determination of the free-energy change for repair of a DNA phosphodiester bond. J. Biol. Chem. 2000; 275:15828–15831. [DOI] [PubMed] [Google Scholar]
- 43. Yang S.W., Chan J.Y.. Analysis of the formation of AMP-DNA intermediate and the successive reaction by human DNA ligases I and II. J. Biol. Chem. 1992; 267:8117–8122. [PubMed] [Google Scholar]
- 44. Cağlayan M., Batra V.K., Sassa A., Prasad R., Wilson S.H.. Role of polymerase β in complementing aprataxin deficiency during abasic-site base excision repair. Nat. Struct. Mol. Biol. 2014; 21:497–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Çağlayan M., Horton J.K., Prasad R., Wilson S.H.. Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res. 2015; 43:2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Beard W.A., Wilson S.H.. Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 1995; 262:98–107. [DOI] [PubMed] [Google Scholar]
- 47. Howes T.R., Tomkinson A.E.. DNA ligase I, the replicative DNA ligase. Subcell. Biochem. 2012; 62:327–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bebenek K., Pedersen L.C., Kunkel T.A.. Replication infidelity via a mismatch with Watson-Crick geometry. Proc. Natl. Acad Sci. U.S.A. 2011; 108:1862–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang W., Hellinga H.W., Beese L.S.. Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc. Natl. Acad Sci. U.S.A. 2011; 108:17644–17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koag M.C., Lee S.. Insights into the effect of minor groove interactions and metal cofactors on mutagenic replication by human DNA polymerase β. Biochem. J. 2018; 475:571–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ahn J., Werneburg B.G., Tsai M.D.. DNA polymerase β: structure-fidelity relationship from Pre-steady-state kinetic analyses of all possible correct and incorrect base pairs for wild type and R283A mutant. Biochemistry. 1997; 36:1100–1107. [DOI] [PubMed] [Google Scholar]
- 52. Morales J.C., Kool E.T.. Minor groove interactions between polymerase and DNA: More essential to replication than Watson-Crick hydrogen bonds. J. Am. Chem. Soc. 1999; 121:2323–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Osheroff W.P., Beard W.A., Yin S., Wilson S.H., Kunkel T.A.. Minor groove interactions at the DNA polymerase β active site modulate single-base deletion error rates. J. Biol. Chem. 2000; 275:28033–28038. [DOI] [PubMed] [Google Scholar]
- 54. Prasad R., Singhal R.K., Srivastava D.K., Molina J.T., Tomkinson A.E., Wilson S.H.. Specific interaction of DNA polymerase beta and DNA ligase I in a multiprotein base excision repair complex from bovine testis. J. Biol. Chem. 1996; 271:16000–16007. [DOI] [PubMed] [Google Scholar]
- 55. Dimitriadis E.K., Prasad R., Vaske M.K., Chen L., Tomkinson A.L., Lewis M.S., Wilson S.H.. Thermodynamics of human DNA ligase I trimerization and association with DNA polymerase beta. J. Biol. Chem. 1998; 273:20540–20550. [DOI] [PubMed] [Google Scholar]
- 56. Prigent C., Satoh M.S., Daly G., Barnes D.E., Lindahl T.. Aberrant DNA repair and DNA replication due to an inherited enzymatic defect in human DNA ligase I. Mol. Cell Biol. 1994; 14:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pascal J.M., O’Brien P.J., Tomkinson A.E., Ellenberger T.. Human DNA ligase I completely encircles and partially unwinds nicked DNA. Nature. 2004; 432:473–478. [DOI] [PubMed] [Google Scholar]
- 58. Tumbale P.P., Jurkiw T.J., Schellenberg M.J., Riccio A.A., O’Brien P.J., Williams R.S.. Two-tiered enforcement of high fidelity DNA ligation. Nat. Commun. 2019; 10:5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cotner-Gohara E., Kim K., Hammel M., Tainer J.A., Tomkinson A.E., Ellenberger T.. Human DNA ligase III recognizes DNA ends by dynamic switching between two DNA-bound states. Biochemistry. 2010; 49:6165–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Conlin M.P., Reid D.A., Small G.W., Chang H.H., Watanabe G., Lieber M.R., Ramsden D.A., Rothenberg E.. DNA Ligase IV guides end-processing choice during nonhomologous end joining. Cell Rep. 2017; 20:2810–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ochi T., Gu X., Blundell T.L.. Structure of the catalytic region of DNA ligase IV in complex with an Artemis fragment sheds light on double-strand break repair. Structure. 2013; 21:672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ochi T., Wu Q., Chirgadze D.Y., Grossmann J.G., Bolanos-Garcia V.M., Blundell T.L.. Structural insights into the role of domain flexibility in human DNA ligase IV. Structure. 2012; 20:1212–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kaminski A., Tumbale P.P., Schellenberg M.J., Williams R.S., Williams J.G., Kunkel T.A., Pedersen L.C., Bebenek K.. Structures of DNA-bound human ligase IV catalytic core reveal insights into substrate binding and catalysis. Nat. Commun. 2018; 9:2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tomkinson A.E., Tappe N.J., Friedberg E.C.. DNA Ligase I from Saccharomycescerevisiae: Physical and biochemical characterization of the CDC9 gene product. Biochemistry. 1992; 31:11762–11771. [DOI] [PubMed] [Google Scholar]
- 65. Shi k., Bohl T.E., Park J., Zasada A., Malik S., Banerjee S., Tran V., Li N., Yin Z., Kurniawan F. et al.. T4 DNA ligase structure reveals a prototypical ATP-dependent ligase with a unique mode of sliding clamp interaction. Nuc. Acids Res. 2018; 46:10474–10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bogenhagen D.F., Pinz K.G.. The action of DNA ligase at abasic sites in DNA. J. Biol. Chem. 1998; 273:7888–7893. [DOI] [PubMed] [Google Scholar]
- 67. Nishida H., Kiyonari S., Ishino Y., Morikawa K.. The closed structure of an archaeal DNA ligase from Pyrococcus furiosus. J. Mol. Biol. 2006; 360:956–967. [DOI] [PubMed] [Google Scholar]
- 68. Chen Y., Liu H., Yang C., Gao Y., Yu X., Chen X., Cui R., Zheng L., Li S., Li X. et al.. Structure of the error-prone DNA ligase of African swine fever virus identifies critical active site residues. Nat. Commun. 2019; 10:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.