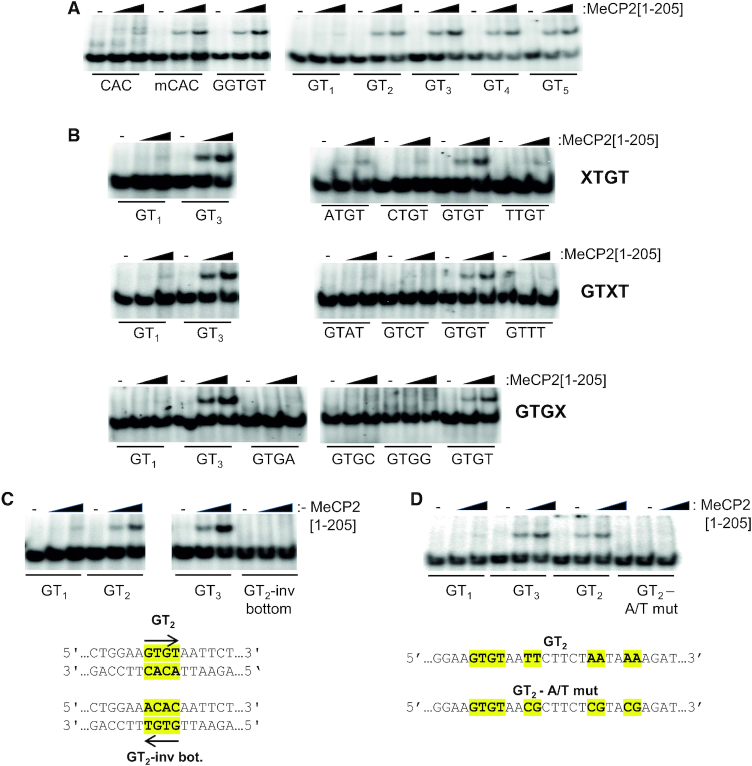
Figure 2.
DNA sequence determinants of cytosine methylation-independent binding by MeCP2. (A) EMSAs using varying amounts of MeCP2[1–205] or no protein (−) with double-stranded oligonucleotide probes containing CAC, mCAC, GGTGT, GT1, GT2, GT3, GT4 or GT5 (see Table 1) to assess the influence of GT-length on binding. Note that CAC is the complement of GTG, indicating that this trinucleotide motif does not bind MeCP2[1–205]. (B) EMSAs in which MeCP2[1–205] was incubated with oligonucleotide probes altered at either the first (XTGT), third (GTXT), or fourth (GTGX) positions in the presence of varying amounts of MeCP2[1–205] or no protein (−). Probes containing GT1 or GT3 were used as negative and positive controls, respectively. (C) Determination of the effect of DNA sequences flanking GTGT binding to MeCP2. EMSAs using varying amounts of MeCP2[1–205] or no protein (−) with probes containing GT1, GT2, GT3 or GT2-inv bot (GT2-inverted indicated by yellow highlighting and inverted arrow). (D) The effect of altering the base composition of the 3′-AT-flank adjacent to GTGT was assayed by EMSAs using varying amounts of MeCP2[1–205] or no protein (−) with probes containing GT1, GT2, GT3 or GT2-A/T mut in which the TT-, AA- and AA-dinucleotides 3′-adjacent to GTGT were substituted by CG (yellow highlight).