Figure 1.
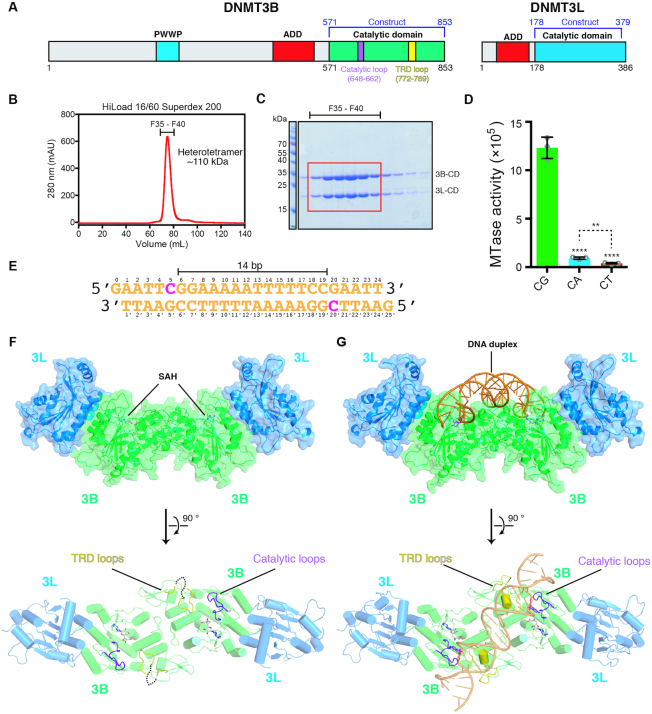
Crystal structures of the catalytic domain of DNMT3B–3L complex bound with and without DNA. (A) Domain structures of DNMT3B and DNMT3L. (B) Catalytic domains of human DNMT3B (residues 571–853) and DNMT3L (residues 178–379) were expressed and co-purified as a heterotetramer with a molecular mass of ∼110 kDa. (C) The co-purified recombinant DNMT3B–3L complex exhibited high homogeneity, as revealed by sodium dodecylsulfate–polyacrylamide gel electrophoresis. (D) Activities of recombinant DNMT3B–3L for CpG, CpA and CpT methylation, as measured by methyltransferase-Glo assays (in luminescence, RLU, n = 3; error bars denote standard deviation (SD); **P < 0.01, ****P < 0.0001). Statistical significance (P-values) was determined by two-tailed Student's t-test. (E) The palindromic DNA duplex containing two CpGpG sites was used for co-crystallization with DNMT3B–3L. (F, G) Crystal structures of DNMT3B–3L complex bound without and with DNA revealing two CpGpG sites bound in the active sites of the DNMT3B dimer (in green). The two SAH molecules are represented by a stick model, catalytic loops are shown in purple and target recognition domain (TRD) loops are displayed in yellow.