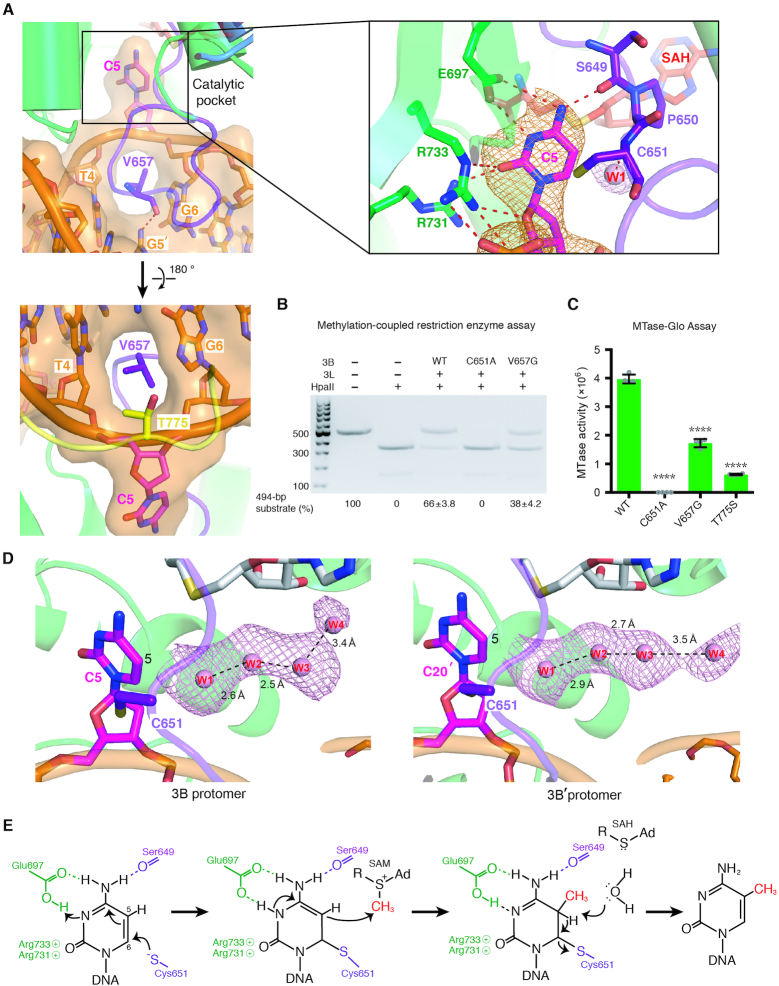
Figure 3.
Specific recognition of the cytosine in CpG sites by DNMT3B. (A) The cytosine (C5) in CpG sites is flipped out of the DNA helix by the catalytic loop. The enlarged panel (upper right) reveals the hydrogen-bond network surrounding the flipped-out cytosine. The omit electron density maps (Fo − Fc) of cytosine (orange) and the nearby water molecule (pink) are contoured at the 4.5σ level. (B) Methyltransferase activities of DNMT3B–3L and the C651A and V657G mutants were estimated by methylation-coupled restriction enzyme HpaII cleavage assays. (C) Methylation activities of wild-type DNMT3B–3L (WT), and the C651A, V657G and T775S mutants were measured by methyltransferase-Glo assays (in luminescence, RLU, n = 4; error bars denote SD; ****P < 0.0001). Statistical significance (P-values) was determined by two-tailed Student's t-test. (D) A water channel located close to the C5 atom of the cytosine of the CpG sites was identified in the two DNMT3B protomers. Cytosines of the CpG sites are shown as magenta sticks, whereas water molecules in the proton wire water channel are shown as pink spheres. The omit electron density maps (Fo − Fc) of water molecules (pink) are contoured at the 2σ level. (E) Proposed working mechanism for methylation by DNMT3B of the cytosine at the C5 position.