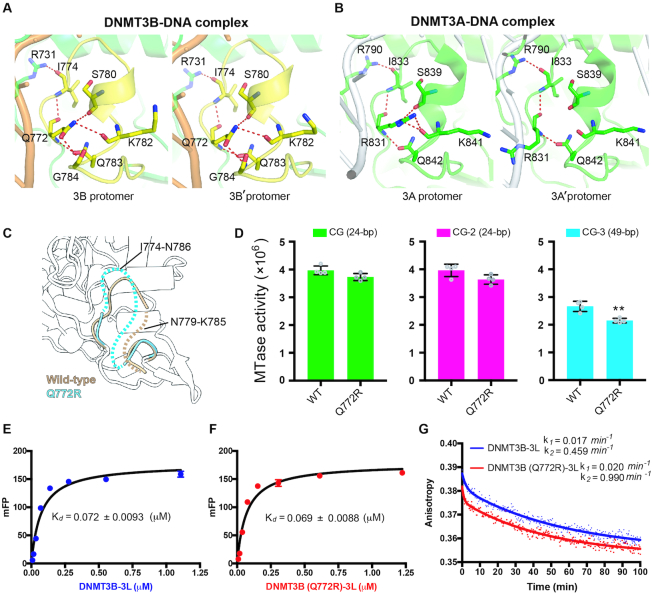
Figure 5.
DNMT3B methylation activity is regulated by TRD loop flexibility. (A) Hydrogen-bond networks mediated by residue Q722 stabilize the TRD loop in both protomers of the DNMT3B dimer in the DNMT3B–3L–DNA complex. (B) Only one complete hydrogen-bond network mediated by residue R831 stabilizes the TRD loop in one of the two protomers of the DNMT3A dimer in the DNMT3A–3L–DNA complex. (C) Superimposition of the crystal structures of wild-type DNMT3B–3L complex (ii) and the DNMT3B (Q772R)–3L complex. Disordered regions in the TRD loop of wild-type DNMT3B–3L (ii) (residues 779–785) and the DNMT3B (Q772R)–3L (residues 774–786) complex are represented by dotted brown or cyan lines, respectively. (D) Methylation activities of DNMT3B–3L and the DNMT3B (Q772R)–3L mutant for DNA of different lengths, as measured by methyltransferase-Glo assays (in luminescence, RLU, n = 4; error bars denote SD; **P < 0.01). Statistical significance (P-values) was determined by two-tailed Student's t-test. (E, F) Dissociation constants (Kd) of DNMT3B–3L and DNMT3B (Q772R)–3L complexes upon binding with the 49-bp DNA (CG-3), as measured by fluorescence polarization (n = 3). (G) Dissociation rate constants (k1 and k2) for DNMT3B–3L and DNMT3B (Q772R)–3L complexes, as determined by measuring the decreasing signal of fluorescence anisotropy of FAM-labeled CG-3 using unlabeled DNA as competitors.