Abstract
During non-homologous DNA end joining (NHEJ), bringing two broken dsDNA ends into proximity is an essential prerequisite for ligation by XRCC4:Ligase IV (X4L4). This physical juxtaposition of DNA ends is called NHEJ synapsis. In addition to the key NHEJ synapsis proteins, Ku, X4L4, and XLF, it has been suggested that DNA polymerase mu (pol μ) may also align two dsDNA ends into close proximity for synthesis. Here, we directly observe the NHEJ synapsis by pol μ using a single molecule FRET (smFRET) assay where we can measure the duration of the synapsis. The results show that pol μ alone can mediate efficient NHEJ synapsis of 3′ overhangs that have at least 1 nt microhomology. The abundant Ku protein in cells limits the accessibility of pol μ to DNA ends with overhangs. But X4L4 can largely reverse the Ku inhibition, perhaps by pushing the Ku inward to expose the overhang for NHEJ synapsis. Based on these studies, the mechanistic flexibility known to exist at other steps of NHEJ is now also apparent for the NHEJ synapsis step.
INTRODUCTION
Unrepaired DNA double-strand breaks (DSB) lead to potentially dangerous chromosome rearrangements. Two major pathways, and several less essential pathways, have evolved to repair DSBs in cells (1,2). Homologous recombination (HR) plays roles in late S and G2 phases of the cell cycle and relies on homologous sequences as the template. Non-homologous end joining (NHEJ) repairs DSBs throughout the cell cycle. NHEJ can directly rejoin the broken DNA ends with little or no homology. NHEJ is more error-prone than HR because it does not restore the original DNA sequence, and therefore leaves an ‘information scar’ (3). Because cells are in a nondividing stage for most of their lifetime, NHEJ is the predominant pathway to repair DSBs within cells. Several proteins are required for NHEJ, depending on the complexity of the broken ends (4). These include Ku, DNA-PKcs, a nuclease (Artemis), polymerases (pol μ, pol λ and TdT), and a ligase complex (XRCC4:LigaseIV:XLF) (5). The variable requirement of these proteins for the final ligation step reflects the mechanistic flexibility of NHEJ. During NHEJ, the two broken dsDNA ends must first be brought into a close proximity - a step called NHEJ synapsis, before their final covalent ligation. Key NHEJ proteins including Ku, XRCC4:Ligase IV (X4L4) and XLF can mediate the formation of a close NHEJ synaptic complex, in which the two dsDNA are aligned in a close-contact end-to-end configuration (6–9).
DNA polymerase mu (pol μ) belongs to the Pol X family polymerase (10). It was initially identified as a homologous protein to TdT, and like TdT, pol μ also displays intrinsic terminal nucleotidyltransferase activity (11–13). Pol μ and pol λ play roles in NHEJ DSB repair and the NHEJ repair of V(D)J recombination intermediates (12,14–16). The gap fill-in activity of the NHEJ polymerases can minimize loss of genetic material and hence reduce the nucleotide deletion within the junction of repair products (12). Deficiency of pol μ in mouse results in shorter immunoglobulin light chain junctions (16,17).
Pol μ contains a BRCT domain, a phosphate binding pocket within the 8 kDa domain, a disordered loop 1 structure, and a polymerase core domain (18). The BRCT domain was reported to interact with other NHEJ proteins, Ku and X4L4 as well as DNA (14,15,19). The interaction between the BRCT domain and DNA could promote the binding of pol μ to DNA and enhance the efficiency of primer extension (19). The 5′ to 3′ polarity of polymerase synthesis is used here to name the upstream DNA end, which serves as a primer for synthesis at its 3′OH, and to contrast it with the downstream DNA end, which provides a template at which pol μ synthesizes across the DSB junction (19–22). The binding of the 5′-P group on the downstream DNA end could also stabilize the pol μ:DNA complex and increase the synthesis efficiency (19). The loop 1 structure of pol μ is disordered in the solved gapped DNA–pol μ structure (21). The loop 1 domain was reported to reposition after the binding of the downstream template and is critical for the template-dependent synthesis of the noncomplementary overhang (21,23). Pol μ has almost the same capability to incorporate ribonucleotides as well as deoxyribonucleotides opposite the deoxyribonucleotide template (20,24,25). The incorporation of ribonucleotides appears to promote NHEJ repair efficiency in cells (26).
Pol μ can add nucleotides at the upstream primer end in a template-dependent or a template-independent manner. The template-independent nucleotide addition (N addition), also known as creative synthesis, is carried out by its terminal transferase-like activity (12,13,27). For the template-dependent synthesis, the two overhangs must be initially aligned into a sufficiently close proximity to make the template on the downstream DNA end available. Pol μ can then fill in the gaps between the upstream and downstream DNA (the primer and the template) ends after alignment of the two overhangs.
The polarity and any microhomology shared between the upstream and downstream DNA ends determines the nature of the contribution of pol X polymerases to NHEJ joining. For ends with 5′ overhangs, these are likely filled to convert them to blunt ends. When the upstream and downstream DNA ends share complementarity in 3′ overhangs, pol μ can use the downstream 3′ overhang as a template to catalyze template-dependent synthesis across the transiently annealed junction. If the two broken duplex ends have no complementary nucleotides, pol μ can carry out template-dependent synthesis across a discontinuous template strand (12,27,28). In this activity, pol μ, preferentially in trans, selects the nucleotide complementary to the one on the downstream noncomplementary 3′ overhang. Both of these forms of synthesis indicate that pol μ can mediate or participate in mediating at least transient NHEJ synapsis of two dsDNA ends, but further details were unclear (29,30).
The possibility of NHEJ synapsis by pol μ has also been examined by a pol μ mediated pull-down assay (31) and a gel shift assay showing a putative NHEJ synaptic complex on gels (27,28,32). No direct evidence has been observed to show that pol μ alone can bring two dsDNA into a close NHEJ synaptic state for a defined duration, and the pull-down and gel shift assays could not confirm if the DNA substrates are aligned in a physiological configuration suitable for synthesis and ligation. Importantly, different studies have shown a variable requirement of other NHEJ key proteins to align the two noncomplementary overhangs for pol μ synthesis (12,28–30,32). The requirement of other NHEJ key proteins for pol μ to align two overhangs may be dependent on the sequence and length of the overhangs (28–30,32).
The crystal structure of the X family polymerase TdT DSB synaptic complex has been reported (33). Efforts have been made to solve the structure of the pol μ DSB synaptic complex. A comparable complex has been reported recently using a TdT chimera of pol μ (23). The TdT chimera is grafted with an extended Loop 1 from pol μ and shown to recapitulate the known functional properties of pol μ. An atomic model of the NHEJ synapsis induced by pol μ can be derived from the structure of the TdT chimera complex. The derived model structure, which uses the crystal 6GO7 PDB as a template, contains pol μ engaged in an NHEJ synapsis with one micro-homology base-pair and one nascent base-pair, and it is found to be stable in molecular dynamic simulations. The derived model could help us understand the behavior of pol μ in NHEJ synapsis, though the TdT chimera cannot completely represent the native pol μ, and no direct experimental evidence has existed to confirm that model.
Here, we provide the first single molecule FRET (smFRET) study to directly monitor the dynamics of NHEJ synapsis by pol μ and investigate the effect of other key NHEJ factors on the pol μ mediated NHEJ synapsis. We find that pol μ alone can carry out NHEJ synapsis of two overhangs sharing at least one base pair of 3′ overhang microhomology. We show that the two duplexes within the NHEJ synaptic complex are aligned in a physiological configuration. We show that Ku protein inhibits the pol μ synapsis if Ku first occupies the DNA end. We show that the X4L4 protein can reverse the inhibition perhaps by pushing the Ku protein inward to expose the overhang for pol μ synapsis. The NHEJ synapsis observed here by pol μ provides an independent pathway to synapsis from that observed with other NHEJ proteins (Ku, X4L4, XLF), and thus demonstrates that the synapsis step is as flexible as other NHEJ steps (6).
MATERIALS AND METHODS
DNA probes
The ssDNA oligonucleotides (oligos) used in this study are made by IDT, and sequences of these oligos are listed in Supplementary Table 1. The Cy3/Cy5-labeled oligos are purified by IDT using HPLC method and directly used without further purification. All other oligos are purified using urea-denaturing PAGE. The duplex DNA molecules are labeled with Cy3 (donor) or Cy5 (acceptor) located 4 bp from the terminus. The cyanine dyes (Cy3 and Cy5) are incorporated into the sugar-phosphate backbone using phosphoramidite chemistry (34). The cyanine dye within the duplex backbone does not significantly affect Ku binding to the DNA duplex. We ran a gel shift assay to estimate the equilibrium dissociation constant (KD) of Ku binding to a 25 bp duplex. The DNA duplexes are BZ91/107 and BZ57/109 (BZ57: 5′-GAT G /iCy3/CC TCC AAG GTC GAC GAT GCA G-3′, BZ109: 5′-CTG C /iCy3/ AT CGT CGA CCT TGG AGG CAT C-3′, BZ91: 5′-GAT GCC TCC AAG GTC GAC GAT GCA G-3′, BZ107: 5′-CTG CAT CGT CGA CCT TGG AGG CAT C-3′). The estimated KD values are 0.086 and 0.11 nM for Ku binding to the 25 bp duplexes without (BZ91/107) and with Cy3 dyes located 4 bp away from both 5′ ends of the duplex (BZ57/109), respectively. The upstream duplexes immobilized on the slide surface are 5′OH/P-BZ15/HC123, 5′OH-BZ15/HC122 and 5′P-BZ14/HC123. They were obtained by annealing corresponding oligos in a buffer of 20 mM Tris–HCl, 100 mM NaCl, pH 8.0. A biotin at the 5′ end of Oligo HC122 and HC123 is used for immobilization onto the slide surface. The incoming downstream duplex has a loop structure at one end that prevents the synapsis reaction from occurring at that end of the duplex. The ‘looped’ duplex (loop-DNA) is made by ligation of a stem-loop structured BZ24 with an annealed Cy3/Cy5-labeled short duplex using T4 DNA ligase (Millipore Sigma, 10716359001) at 15°C overnight. The short duplexes for loop-DNA preparation include 5′OH / P-BZ35/BZ36, 5′P-BZ35/83, 5′P-BZ35/84 and 5′P-BZ85/36. After ligation, the intended ligation product, the looped duplex, is then isolated from input short duplexes and self-ligated products using a 12% native PAGE.
Protein expression and purification
Pol μ full-length (FL) and the ΔBRCT deleted version (pol μ-ΔBRCT, aa133–aa494) are the same purified batches as used in a previous study (15). Ku70/80 heterodimer (Ku) and X4L4 are also the same purified batches as used in a previous study (6). All the recombinant proteins are documented to be functional in binding and NHEJ assays. The protein concentrations were measured using SDS-PAGE gel and using BSA as a standard.
Slide and coverslip modification
Slides and coverslips for imaging are modified as previously described (6). Briefly, slides and coverslips are sequentially treated with 4 M NaOH for 30 min and ‘piranha’ solution (3 volumes H2SO4 (98% v/v) to 1 volume H2O2 (30% v/v) for another 30 min. The slides and coverslips are thoroughly washed with distilled water after each step and lastly washed twice with 100% methanol. The slides and coverslips then remain in HPLC-grade methanol until the next step. Methanol solution containing 2% (v/v) (3-aminopropyltriethoxy)silane (Millipore Sigma, 440140) and 5% (v/v) acetic acid is then used to silanize the slide and coverslip. After washing with water, and then drying, slides and coverslips are PEGylated by using a viscous solution of mPEG (Laysan Bio, mPEG-SVA-5000) and a mixture of mPEG and Biotin-PEG (Laysan Bio, Biotin-PEG-SVA-5000), respectively. The ratio of mPEG to Biotin-PEG is set to 40:1 for coverslips. PEGylation is done at room temperature in the dark overnight. The slides and coverslips are then thoroughly washed with distilled water, dried and kept at −20°C until use.
Single-molecule imaging
The microscopy system used here is the same one as used for a previous study (6). Unless otherwise specified, all reactions are carried out at room temperature (22°C) with a buffer containing 20 mM Tris-acetate pH 7.5, 75 mM KAc, 10 mM MgAc2, 3% glycerol, 1 mg/ml BSA, 2 mM DTT, 0.25% Tween 20, 100 μM nucleotide, 2 mM Trolox (Millipore Sigma, 238813) and an oxygen scavenger system [0.8% (w/v) d-glucose (RPI Research Products International, G32040-5000) and 1× gloxy mix consisting of 165 U/ml glucose oxidase (Millipore Sigma, G2133–50KU), and 2170 U/ml catalase (Millipore Sigma, C3556)]. For all reactions except those in Supplementary Figure S1B and C, 250 pM 5′P/OH-BZ15/HC123 or 5′OH-BZ15/HC122 or 5′P-BZ15/BZ82 dsDNA as specified on the figures is immobilized on the coverslip surface via the biotin-neutravidin-biotin strategy, and 10 nM Cy3-labeled loop–DNA is used in the solution. For reactions in Supplementary Figure S1B and C, 10 pM 5′P-BZ15/HC123 and 5′P-BZ14/HC123 are immobilized on the surface, respectively. 10 nM Cy3 labeled 5′P-(BZ24+BZ35/BZ36) dsDNA and 10 nM Cy5 labeled 5′P-(BZ24+BZ85/BZ36) dsDNA are used in the reactions of Supplementary Figure S1B and C, respectively. The final protein concentrations applied in the study are as follows unless otherwise specified: 50 nM Ku, 50 nM X4L4, 50 nM pol μ and 50 nM pol μ-ΔBRCT. To test the ligatability of the overhangs within the NHEJ synaptic complex (Figures 1F, 6D and E; Supplementary Figure S2), duplexes with proteins as specified in the figures are incubated for 30 min at room temperature. After that, the reaction chamber on the slide is imaged at three randomly chosen regions. Then 2 M NaCl in T50 buffer (10 mM Tris–HCl pH 8.0, 50 mM NaCl) flows through the reaction chamber to remove any noncovalent associations of the duplexes. The reaction chamber is then imaged in the reaction buffer at the three regions.
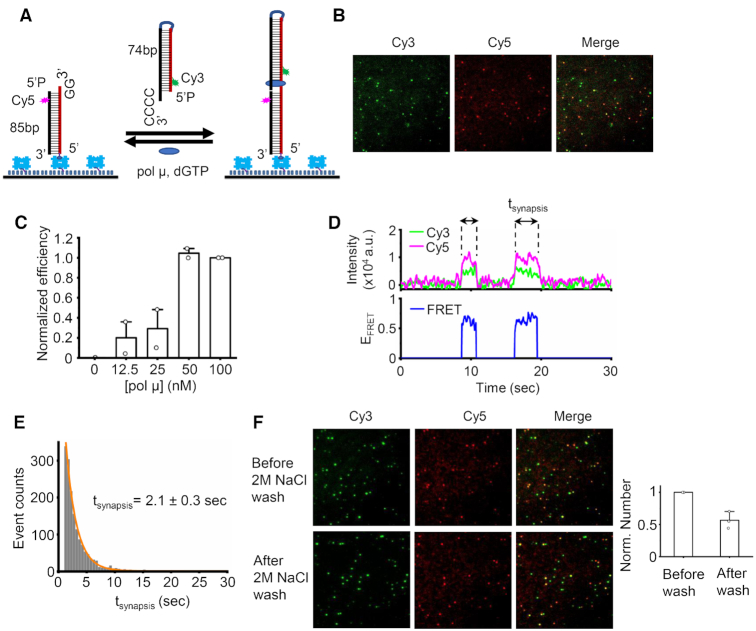
Figure 1.
Human pol μ can mediate efficient NHEJ synapsis. (A) Schematic of smFRET assay for pol μ mediated synapsis. An 85 bp duplex with a 3′ GG overhang and a Cy5 dye (acceptor) located 4 bp away from the end was immobilized on the surface of the imaging coverslip. The 74 bp incoming dsDNA with a 3′ CCCC overhang and a Cy3 dye (donor) located 4 bp away from the end was added to the reaction via a 20 μl injected solution that also contained specified concentrations of pol μ and 100 μM dGTP. Each duplex has a terminal 5′-P group. (B) Representative images of Cy3 channel, Cy5 channel, and overlay of two channels for synapsis mediated by 50 nM pol μ. (C) Normalized NHEJ synapsis efficiency mediated by varied amount of pol μ. Data are represented as the mean ± SD of two independent replicates. (D) One of the single molecule time traces of donor (green) intensity, acceptor (magenta) intensity and corresponding EFRET values (blue) for NHEJ synapsis mediated by 50 nM pol μ. (E) Histogram and corresponding single exponential fit of NHEJ synapsis time of complex exhibiting a FRET of ∼0.6. At least 1000 NHEJ synapsis events were included. NHEJ synapsis time shown on the graph is represented as the mean ± SD of four replicates. (F) Representative images of Cy3 channel, Cy5 channel, and overlay of two channels for NHEJ synaptic molecules before/after 2 M NaCl wash. The bar graph represents the quantified results. 50 nM pol μ, 50 nM X4L4 and 100 μM dGTP were included in the reactions. We used 2 M NaCl to remove any noncovalent associations of the overhangs, which leaves only the covalently ligated molecules remaining.
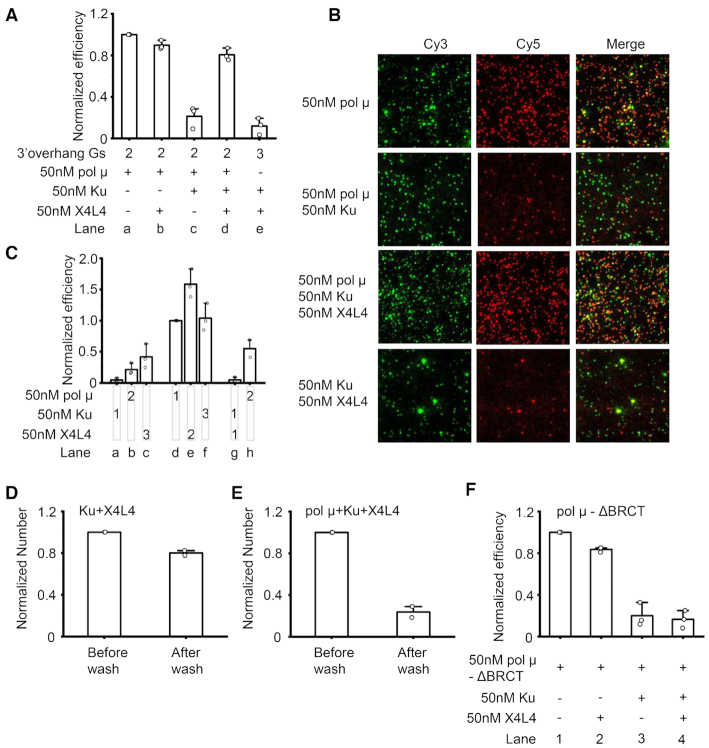
Figure 6.
X4L4 can largely reverse Ku inhibition of pol μ-mediated synapsis. (A) Normalized efficiency of NHEJ synapsis mediated by different combinations of 50 nM pol μ, 50 nM Ku and 50 nM X4L4. The incoming duplex, proteins, and 100 μM ddGTP were first mixed well, then injected into the reaction chamber. The DNA duplexes have 3′ GG and 3′ CCCC overhangs unless otherwise specified and have terminal 5′-OH groups. The numbers 2 and 3 on the graph represent two Gs and three Gs, respectively at the immobilized duplex 3′ overhang. Data are represented as the mean ± SD of three independent replicates. The use of the three Gs overhang substrate for Ku plus X4L4 synapsis is to completely exclude the contribution of Ku plus X4L4 to the synapsis efficiency. (B) Representative images of Cy3 channel, Cy5 channel, and overlay of the two channels for NHEJ synapsis mediated by different combinations of NHEJ proteins shown in (A). (C) Effect of the order of NHEJ protein addition on synapsis efficiency. 10 nM incoming dsDNA and 100 μM ddGTP were first mixed and added into the reaction chamber. 50 nM pol μ, 50 nM X4L4, and 50 nM Ku were then sequentially added into the chambers as specified. 1 represents the first addition, 2 represents the second addition, 3 represents the third addition. The DNA duplexes have 3′ GG and 3′ CCCC overhangs and have terminal 5′-OH groups. Data are represented as the mean ± SD of at least two independent replicates. (D, E) Normalized amount of NHEJ synaptic molecules before/after 2M NaCl wash. 50 nM Ku and 50 nM X4L4 (D), or 50 nM Ku, 50 nM X4L4 and 50 nM pol μ (E) were included in the reactions. No nucleotides were present in the reactions. The DNA duplexes have 3′ GG and 3′ CCCC overhangs and have terminal 5′-P groups. Data are represented as the mean ± SD of two independent replicates. (F) Normalized efficiency of NHEJ synapsis mediated by different combinations of 50 nM pol μ ΔBRCT, 50 nM Ku, and 50 nM X4L4. The incoming duplex, proteins, and 100 μM ddGTP were first mixed well, then injected into the reaction chamber. The DNA duplexes have 3′ GG and 3′ CCCC overhangs and have terminal 5′-OH groups. Data are represented as the mean ± SD of three independent replicates.
Unless otherwise specified, proteins and looped dsDNA are added stepwise into the reaction buffer and quickly mixed. The reaction mixture is then immediately injected into the imaging chamber, which is made with a slide and a coverslip with the upstream duplex already immobilized. Images are captured immediately after sample injection or after incubation for certain times as specified on the figures. For reactions in which the order of protein addition is adjusted (Figures 5B–D and 6C; Supplementary Figure S7A–C), the downstream duplex and nucleotide are first mixed well in the reaction buffer and added into the reaction chamber. Specific protein or protein combinations are then separately added into the reaction mixture and gently mixed. The sample is imaged after the addition of each protein/protein combination and an incubation for 4 min. Three movies of 30 s duration are captured using an exposure time of 200 ms for each protein/protein combination addition. For all other reactions, CCD camera exposure time is set at 50 ms. At least 3 movies are captured for each sample, and each movie lasts ∼1 min. At least two independent samples are analyzed for each condition. A 514 nm laser is always used to excite the FRET pairs during each ROI imaging. Cy5 dye cannot be directly excited by the 514 nm laser. It can only become bright when the Cy5 dye obtains the energy transferred from the excited Cy3 dye. The emission fluorescence from Cy3 and Cy5 is split into two perpendicularly mounted Ixon+897 Ultra EMCCD cameras (Andor) using a dichroic mirror (FF640-FDi01-25x36, Semrock). After capturing each ROI using 514 nm laser, the same ROI is directly excited by a 647 nm laser to confirm the homogeneity of the immobilized Cy5-dsDNA.
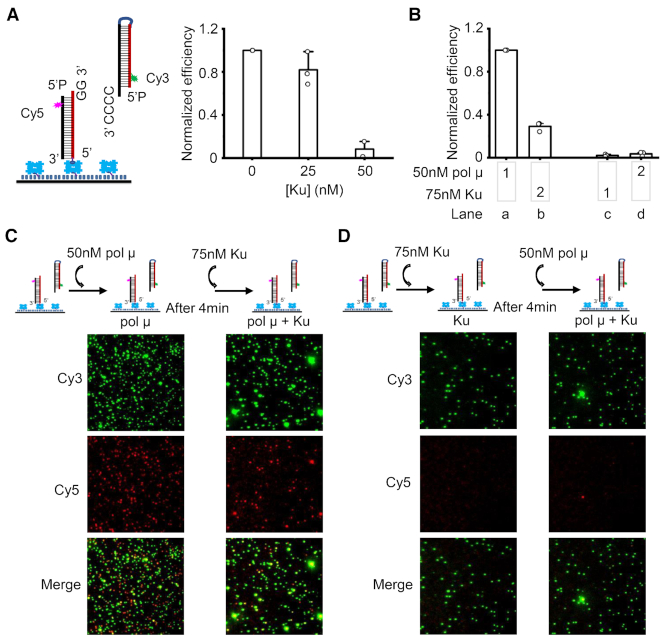
Figure 5.
Ku binding inhibits pol μ mediated synapsis. (A) Ku concentration-dependent inhibition of pol μ mediated synapsis. 50 nM pol μ, 100 μM dGTP, specified concentrations of Ku, and 10 nM incoming duplex were first mixed well, then injected into the reaction chamber. Both duplexes have terminal 5′-P groups. Data are represented as the mean ± SD of at least two independent replicates. (B) Effect of the order of Ku addition on NHEJ synapsis efficiency. 10 nM incoming dsDNA and 100 μM ddGTP were first mixed and added into the reaction chamber. 50 nM pol μ and 75 nM Ku were then sequentially added into the chambers as specified. 1 represents the first addition, 2 represents the second addition. Both duplexes have terminal 5′-OH groups. Data are represented as the mean ± SD of three independent replicates. (C, D) Representative images of Cy3 channel, Cy5 channel, and overlay of the two channels for NHEJ synapsis after certain protein addition. The quantified synapsis efficiency is depicted in (B).
smFRET data analysis
All FRET pairs and corresponding single molecule trajectories are searched and extracted using the iSMS v2.01 software (35,36) written in MATLAB. The data analysis method is similar to that described in our previous study (6). Briefly, the FRET pairs are searched by scanning every 10 frames of the movie. The final FRET pair number is obtained after the visual identification of all the trajectories. Since the slide surface is not perfectly blocked, solution Cy3-dsDNA can non-specifically adsorb onto the surface, which results in some spots on the images due to Cy3 only (called ‘Cy3-only spots). Only the molecules with paired Cy3 and Cy5 signals were selected. Since >90% of the identified molecules only exhibited one synapsis event within the detection time (Supplementary Figure S3B), the total synapsis efficiency is calculated from the number of final molecules identified (which have FRET pairs) instead of the synapsis events. Because of the slide-to-slide variance, the synapsis efficiency is always normalized for the number of molecules obtained from the reaction of 50 nM pol μ alone conducted on the same slide, unless otherwise specified.
All identified trajectories are then exported, and both donor and acceptor raw intensity data are smoothed by averaging the neighboring five data points. The data is further analyzed using programs written in MATLAB as described previously (7,37). All the molecules from each specific condition are combined and used for normalized EFRET histograms. The photobleached portions of each trajectory are first removed, and the first 10 data points of each trajectory are included in the histograms.
Synapsis time analysis
The synapsis time is calculated based on the Cy3 signal of the identified FRET molecule. All the dwell time histograms and the corresponding exponential fits only correspond to the synapsis events with both beginning and end times within the detection time window. All the dwell time histograms except that stimulated by ddGTP are fitted by a single exponential function. The synapsis time histogram stimulated by ddGTP could not be fitted with a single exponential function. It is fitted by a bi-exponential function. The short time of the bi-exponential fit may represent the synaptic complex without nucleotide present.
Quantification and statistical methods
The unpaired, two-tailed t-test is performed using Microsoft Excel. All other data is compiled in OriginLab Origin 2019.
RESULTS
Human pol μ can mediate efficient NHEJ synapsis
A similar strategy for NHEJ synapsis detection is applied here to monitor the NHEJ synapsis of pol μ, as previously described (6). We first tested if pol μ alone can promote NHEJ synapsis of duplexes with partially complementary 3′ overhangs. One DNA duplex with a 3′ protruding GG is immobilized on the slide surface, and the other DNA duplex with a 3′ CCCC overhang is free in solution and introduced into the reaction chamber together with pol μ plus dGTP (Figure 1A). The amount of immobilized duplex is at least 40-fold less than that of the solution duplex. Therefore, the 3′ CCCC overhang has a much greater probability to serve as a template for pol μ synthesis after any synapsis. The free solution duplex provides the downstream DNA end, as defined earlier. The 3′GG overhang has a much greater probability to serve as the upstream primer end, as defined above. The immobilized upstream duplex has a Cy5 tag as a FRET acceptor. The incoming downstream DNA from the solution is labeled with Cy3 as a FRET donor near the end. The applied 514 nm laser during imaging is specific for Cy3 donor excitation. The Cy5-dsDNA can become bright only when the Cy5 acceptor obtains energy transferred from the excited Cy3 dye. Therefore, no FRET signal can be observed unless the downstream and upstream duplexes are brought together by pol μ to form an NHEJ synaptic complex. The simultaneous appearance of Cy3 and Cy5 signals indicates the interaction of the two DNA ends.
Image results show that some molecules have colocalized Cy3 and Cy5 signals (Figure 1B). As discussed above, the colocalized Cy3 and Cy5 signals suggest NHEJ synapsis occurs. This indicates that pol μ alone can mediate NHEJ synapsis of the two duplexes. The NHEJ synapsis was further confirmed by the simultaneous appearance of Cy3 and Cy5 signals on the typical trajectories (Figure 1D). The synapsis efficiency exhibits a pol μ concentration-dependent increase and reaches a plateau at 50 nM (Figure 1C). This result further demonstrates that the observed NHEJ synapsis is mediated by pol μ. FRET efficiency is distance sensitive. It can reflect the relative distance between the donor and acceptor. Therefore, we then examined the relative positions of the two ends within the synaptic complex by checking the EFRET profile of a single pair of molecules and also the EFRET distribution of all the molecules in a given experiment.
The formed NHEJ synaptic complexes exhibit two different EFRET states; the major complex has a FRET of 0.6, and the FRET of the minor state is 0.3 (Figure 1D; Supplementary Figure S1A and B). The FRET results suggest that the two duplexes within different complexes may have different configurations. By carefully checking the trajectory, we can see that the total intensity of donor and acceptor of the minor 0.3 FRET synaptic complex is almost twice that of the major 0.6 FRET state, while the two states have almost the same level of Cy5 intensity (Supplementary Figure S1A). The result suggests that the synaptic complex with 0.3 FRET contains two Cy3-labeled downstream duplexes (see Supplement). Pol μ exists as a monomer in solution (11), and the solved structures of pol μ:DNA complex accommodate merely one gapped duplex (21,22,38). Therefore, the observed minor amount of three-duplex synaptic complex is likely due to artificial factors. Hereafter, we focus on the NHEJ synaptic complex with the major FRET of 0.6, which contains one upstream duplex and one incoming downstream one, and which has the clearest physiological relevance (see below). The formed NHEJ synaptic complex with 0.6 FRET can last for 2 s (Figure 1E). The increase of the end microhomology (MH) from two nucleotides (nt) MH to 3 nt MH does not further increase the lifetime of the formed synaptic complex (Supplementary Figure S1F).
Previous studies have shown that pol μ prefers to skip the first available template nucleotides and preferentially select the deoxyribonucleotide triphosphate complementary to the template nucleotide close to the recessed 5′ end of the downstream DNA (38,39). Importantly, the EFRET standards obtained from our system suggest that 10–12 bp distance between the Cy3 and Cy5 dyes could result in a FRET of ∼0.6 (Supplementary Figure S1D). The two dyes are both 4 bp away from the ends; therefore, the NHEJ synaptic state with a FRET of 0.6 corresponds to a DSB mimic complex in which the G at the very end of the upstream primer pairs with the second C from the end of downstream template (Supplementary Figure S1E). The DSB synaptic complex is sensitive to the incoming nucleotide. The two overhangs can be ligated in 56% of the synaptic complexes when the reaction condition permits dG nucleotide addition (Figure 1F; Supplementary Figure S2A and B). This indicates that the two overhangs are indeed brought into a close proximity by pol μ for synthesis, followed by ligation. Although pol μ can mediate NHEJ synapsis of the two overhangs in the absence of any nucleotides (see below and Supplementary Figure S2C), the two overhangs cannot be ligated without dG addition to the upstream 3′ end (Supplementary Figure S2C). These results indicate that besides pol μ-mediated synapsis, the pol μ-mediated polymerization is also required for the final ligation of the two overhangs. These results further confirm the configuration of DNA within the formed NHEJ synaptic complex, which shows that a gap exists between the two aligned ends (Supplementary Figure S1E).
Microhomology is required for efficient NHEJ synapsis
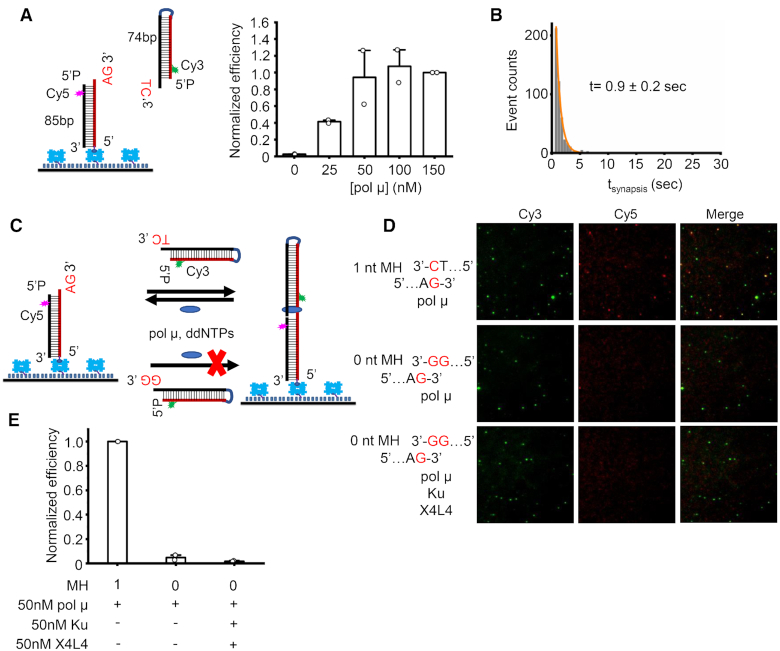
Previous studies suggest that pol μ alone or aided by Ku70/Ku80 (Ku) and XRCC4:LigIV (X4L4) can use the downstream incompatible overhang as the template to carry out template-dependent synthesis (29,30), suggesting that pol μ may participate in bridging two incompatible overhangs. We next tested if pol μ alone can mediate NHEJ synapsis of two duplexes with incompatible overhangs. The upstream duplex has a 3′ AG overhang as the primer. The downstream duplex was designed to either have a 3′ TC overhang which can provide 1 nt microhomology (1 nt MH) for the primer (Figure 2A and Supplementary Figure S3A) or have a 3′GG overhang which is incompatible (0 nt MH) with the primer (Figure 2C). The results show that pol μ alone stimulated by dATP can mediate efficient NHEJ synapsis of the duplexes with 1 nt MH (Figure 2A and Supplementary Figure S3A-D). The synapsis efficiency exhibits a pol μ concentration-dependent increase and reaches a plateau at ∼50 nM (Figure 2A). The NHEJ synaptic complex lasts for 0.9 sec (Figure 2B), which is shorter than that of the complex formed with overhangs having 2 nt microhomologies (Figure 1E). This result indicates that overhang microhomology can stabilize and increase the longevity of the formed NHEJ synaptic complex.
Figure 2.
Microhomology is required for efficient NHEJ synapsis. (A) Left panel: Schematic of smFRET assay for NHEJ synapsis of overhangs with 1 nt microhomology. Right panel: Normalized synapsis efficiency of overhangs with 1 nt microhomology mediated by specified amounts of pol μ. Data are represented as the mean ± SD of two independent replicates. 100 μM dATP was included in the reactions. (B) Histogram and corresponding single exponential fit of synapsis time of complex exhibiting a FRET of ∼0.6. The reaction corresponds to that using 50 nM pol μ as shown in (A). At least 190 synapsis events were included. Synapsis time shown on the graph is represented as the mean ± SD of two replicates. (C) Schematic of smFRET assay for NHEJ synapsis of overhangs with or without microhomology (MH). The immobilized duplex has a 3′ AG overhang, the solution duplex has either a TC (1 nt MH) or GG (0 nt MH) overhang. 50 nM pol μ and ddNTPs (100 μM each) in the presence/absence of 50 nM Ku and 50 nM X4L4 were included in the reactions. (D) Representative images of Cy3 channel, Cy5 channel, and overlay of the two channels for NHEJ synapsis of different sets of duplexes. (E) Normalized NHEJ synapsis efficiency for different sets of DNA duplexes. Data are represented as the mean ± SD of at least two independent replicates. MH represents base-pair number of microhomology.
We then tested the NHEJ synapsis of incompatible overhangs. dNTPs (Supplementary Figure S3E–G) or ddNTPs (Figure 2C–E) were present in the reactions for any types of nucleotide addition. Pol μ can mediate efficient synapsis of the overhangs with 1 nt MH, but only rare NHEJ synaptic molecules were observed for the duplexes having incompatible overhangs (Figure 2C–E and Supplementary Figure S3E–G). We also tested whether Ku and X4L4 simultaneously present in the reactions could help the pol μ synapsis of incompatible overhangs, because it was reported that Ku and X4L4 could help pol μ carry out synthesis across the discontinuous template strand using the downstream incompatible overhang (29). Here we focus purely on the synapsis step; therefore, ddNTPs were present in the reactions to prevent any covalent ligations by X4L4 (Figure 2C). The results show that pol μ even with the aid of Ku and X4L4 could not mediate durable synapsis for the incompatible overhangs (Figure 2D and E). The results above highlight that overhang microhomology is important and required for efficient NHEJ synapsis mediated by pol μ.
Recessed 5′ P group at the downstream dsDNA does not have a large effect on pol μ synapsis
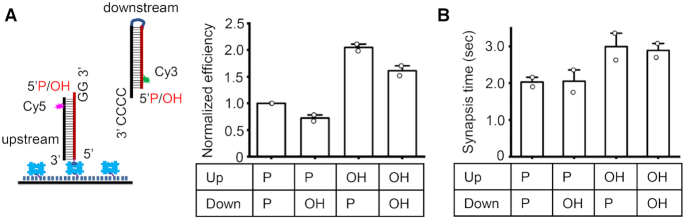
Pol μ has a phosphate binding pocket within an 8 kDa domain (18), which was previously reported to recognize and bind the recessed 5′-P group to stabilize the pol μ:DNA complex (19,23). Recognition of the recessed 5′ P group by pol μ was also suggested to be crucial for its template-directed extension of the primer; that is, the primer strand can use the nucleotides in a second incompatible strand as template for nucleotide addition (19,30). We then examined if the recessed 5′ P group at the downstream duplex end as well as at the upstream DNA end have any effects on the pol μ synapsis. The synapsis efficiency results show that the 5′-P group on the downstream duplex, in contrast to its role in promoting pol μ:DNA complex as previously described (19), does not have a large effect on the physiologically relevant synapsis (Figure 3A and Supplementary Figure S4). The synapsis efficiency only decreases 20–30% when a 5′-OH group is at the downstream end (Figure 3A). Moreover, the 5′-P group has almost no effect on the duration of the formed synaptic complex (Figure 3B).
Figure 3.
Effect of DNA end chemistry on pol μ mediated synapsis. (A) Left panel: DNA duplexes used for the assay. Right panel: Normalized NHEJ synapsis efficiency for DNA duplexes having different end chemistry. The DNA duplex either has a 5′-P group or has a 5′-OH group. 50 nM pol μ and 100 μM dGTP were included in the reactions. Data are represented as the mean ± SD of two independent replicates. (B) Dwell time of NHEJ synaptic complex. Data are represented as mean ± SD of two independent replicates.
In contrast to the downstream 5′-P group, the upstream 5′-P group decreases the synapsis efficiency by 2-fold (Figure 3A) and reduces the duration of the synaptic complex by 40% (Figure 3B). This is compatible with the crystal structure of the TdT chimera recapitulating pol μ functional properties and crystallized with a DNA-ends synapsis (23), which shows that a 5′-P group in the upstream template strand would clash with the protein and prevent the formation of a stable complex. Therefore, in contrast to the end chemistry of the downstream end, the end chemistry of the upstream DNA can affect the pol μ synapsis.
Nucleotides enhance the pol μ synapsis, but are not essential
We also tested if the presence of a nucleotide affects pol μ synapsis. The upstream duplex has a 3′ GG overhang, and the downstream duplex has a 3′CCCC overhang (Figure 4A). Therefore, deoxyguanine triphosphate (dGTP) and dideoxyguanine triphosphate (ddGTP) are the complementary deoxyribonucleotides opposite C. Deoxythymidine triphosphate (dTTP) is used as the noncomplementary deoxyribonucleotide control. Pol μ can also incorporate ribonucleotide opposite the deoxyribonucleotide template (20,24,25), and incorporation of ribonucleotide is suggested to be required for the biological activity of pol μ (26). Therefore, the complementary riboguanine triphosphate (rGTP) was also tested.
Figure 4.
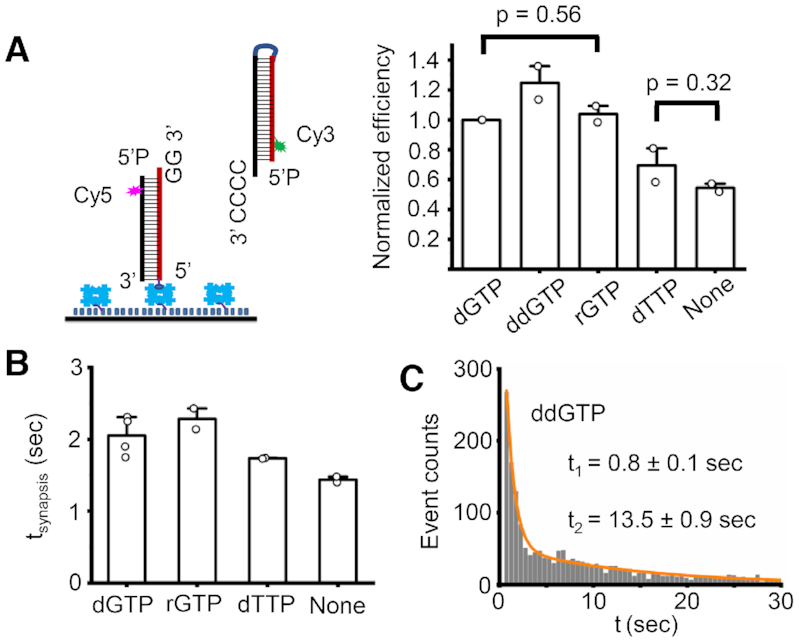
Effect of nucleotide on pol μ mediated synapsis. (A) Normalized efficiency of NHEJ synapsis stimulated by different nucleotides. 50 nM pol μ with or without 100 μM nucleotide were included in the reactions. Data are represented as the mean ± SD of two independent replicates. T-test (unpaired, two-tailed) was applied for p value calculation. (B) Dwell time of NHEJ synaptic complex stimulated by different nucleotides (dGTP, rGTP, dTTP). The synapsis time was obtained from a single exponential fit of the dwell time histogram (Supplementary Figure S5C). Data are represented as the mean ± SD of at least two independent replicates. (C) Histogram and corresponding bi-exponential fit of NHEJ synapsis time stimulated by 100 μM ddGTP. At least 1000 synapsis events were included. Synapsis time shown on the graph is represented as the mean ± SD of two replicates. The short time t1 obtained from the bi-exponential fit may represent the duration of NHEJ synaptic complex formed without any nucleotides present.
We find that pol μ can mediate NHEJ synapsis in the absence of any cofactors (Figure 4A and Supplementary Figure S5A), which indicates that a nucleotide is not required for the synapsis. The non-complementary dTTP does not significantly increase synapsis efficiency (Figure 4A). All the complementary nucleotides (dGTP, ddGTP, and rGTP) enhance the synapsis efficiency by ∼2-fold (Figure 4A and Supplementary Figure S5A). rGTP has almost the same capability as dGTP to stimulate the synapsis, likely because pol μ does not have a preference to recognize different sugar moieties of nucleotides (20,24,25). The tested nucleotides except ddGTP do not have a large effect on the duration of the synaptic complex, compared to that without nucleotide stimulation (Figure 4B and Supplementary Figure S5C), though they can increase the synapsis efficiency. The lifetime of the synaptic complexes, except that stimulated by ddGTP, is ∼2 s. ddGTP clearly stabilizes the synaptic complex to 13 s (Figure 4C and Supplementary Figure S5C). The longer duration of synaptic complex stimulated by ddGTP suggests that the chemistry of the upstream duplex has a large effect on the NHEJ synapsis as described above (Figure 3).
Ku inhibits pol μ mediated synapsis
Key NHEJ proteins were previously reported to mediate NHEJ synapsis of two broken DNA ends (6–9,40). Pol μ can interact with Ku in the presence or absence of X4L4 to form a stable complex on dsDNA, and the BRCT domain of pol μ was reported to be critical for this interaction (14,15). We next wanted to see if the key NHEJ factors, especially Ku and X4L4 would affect the pol μ synapsis. As in the previous experiments for pol μ synapsis, all proteins and the incoming downstream duplex accompanied with nucleotide were first mixed and simultaneously added into the slide chamber on which the upstream duplex had already been immobilized.
The results show that Ku inhibits the pol μ synapsis in a concentration-dependent manner (Figure 5A). Ku at 25 nM concentration does not affect the pol μ mediated synapsis, but Ku at 50 nM concentration decreases the efficiency of pol μ synapsis by ∼90% (Figure 5A). The inhibition of Ku is not due to the interaction between Ku and pol μ. This is clear because the BRCT domain deleted pol μ (pol μ-ΔBRCT), which cannot interact with the Ku protein (15), has similar activity as the full-length (FL) protein to promote synapsis (Supplementary Figure S6); moreover, Ku still inhibits the synapsis mediated by the pol μ-ΔBRCT (Supplementary Figure S6E).
The duplexes used here are 74–85 bp in length. Up to 3 Ku molecules can simultaneously bind to each duplex when sufficient Ku protein is present in the reaction solution (41). When Ku is at a relatively high concentration of 50 nM, the molar ratio of Ku to DNA duplex is around 5:1. This means three Ku molecules can bind to each duplex. The binding of three Ku molecules would occupy the entire duplex. This would limit the accessibility of the overhangs of the duplex to pol μ, which would inhibit the pol μ synapsis. Importantly, 25 nM Ku merely slightly impairs the pol μ synapsis (Figure 5A) because pol μ can still access the duplex overhang, after Ku binds to the duplex. Therefore, these results indicate that Ku occupancy on the DNA inhibits pol μ synapsis. The inhibition by Ku occupancy was further confirmed by adjusting the order of protein addition (Figure 5B–D). We find that if pol μ is present first in the reaction and mediates synapsis, some of the synaptic complexes (30%) can still remain after Ku addition (Figure 5B, lanes a, b; C). But if Ku is added into the reaction chamber first and occupies the duplex DNA, then pol μ is almost entirely unable to mediate synapsis (Figure 5B, lanes c, d; D). These results imply that binding of Ku to the entire two duplexes can limit the accessibility of DNA overhangs to pol μ, which thus inhibits any pol μ-mediated synapsis.
X4L4 can largely reverse Ku inhibition of pol μ-mediated NHEJ synapsis
Because Ku protein is abundant in cells (5), the inhibition due to Ku could mean that pol μ synapsis is insignificant within cells. We wondered if other NHEJ proteins could reverse the Ku inhibition. Previous studies showed that pol μ aided by Ku and X4L4 could conduct template-dependent synthesis using the noncomplementary template on a second dsDNA, suggesting end bridging is required for pol μ’s function in repair of noncomplementary ends (29). We tested if X4L4 could reverse the Ku inhibition observed here.
We first tested the possibility of such a relief of inhibition by adding all of the proteins, pol μ, Ku and X4L4 together into the reaction. Previous studies suggested that end microhomology can stimulate the formation of the end-to-end configured synaptic state (designated ‘close synaptic’ complex) by Ku plus X4L4, although they mainly promote the formation of the ‘flexible synaptic complex’ in which the two duplexes are laterally aligned (6,7). The results here further indicate Ku plus X4L4 can stimulate the end-to-end close synapsis and longer end microhomology can facilitate this close synapsis (Supplementary Figure S7D). Pol μ can add nucleotides to the upstream 3′ 2G overhang; therefore, the upstream substrate with a 3′ 3G overhang was used to completely exclude the contribution of Ku plus X4L4 to the synapsis efficiency (Figure 6A, lane e). The synapsis efficiency results show that 50 nM Ku obviously inhibits the NHEJ synapsis and reduces the efficiency to 0.2, which is 80% lower than without Ku present in the reaction (Figure 6A, lanes a, c; B). In contrast, the synapsis efficiency rises back to 0.8 when 50 nM X4L4 is simultaneously present in the reaction (Figure 6A, lane d; B). The increase in NHEJ synapsis efficiency is not entirely due to the contribution of the close synapsis mediated by Ku plus X4L4, observed previously (6,7). This is because the efficiency of close synapsis mediated by Ku plus X4L4 is only 0.12 (Figure 6A, lane e; B), which, even considering the efficiency seen with pol μ and Ku (Figure 6A, lane c), is much lower than the efficiency mediated by pol μ, Ku, and X4L4 (Figure 6A, lane d). The results indicate that X4L4 can clearly reverse the Ku inhibition. A previous study suggested that X4L4 binding to Ku-bound DNA could push the Ku inward and that the freely inward translocation of Ku on the DNA is essential for Ku to stimulate the X4L4 activity (42). The reversion of Ku inhibition by X4L4 observed here indeed may be because it can push the end Ku protein inward to expose the overhangs for pol μ synapsis.
The reversion by X4L4 was further confirmed by adjusting the order of protein addition (Figure 6C and Supplementary Figure S7A–C). When X4L4 is added before or with Ku to the reaction, pol μ can mediate efficient NHEJ synapsis (Figure 6C, lanes d–h; Supplementary Figure S7B and C). As in the previous result, the initial addition of 50 nM Ku inhibits pol μ synapsis (Figure 6C, lanes a–b; Supplementary Figure S7A); and the subsequent addition of 50 nM X4L4 partially reverses the inhibition and increases the NHEJ synapsis efficiency 2-fold (Figure 6C, lanes b–c; Supplementary Figure S7A).
To further confirm that the synapsis by the combination of Ku, X4L4 and pol μ is still pol μ-dependent, a new experiment was designed based on the configuration of the two DNA substrates within the close synaptic complex. The results show that 80% substrates within the synaptic complexes formed by Ku plus X4L4 are ligated (Figure 6D), which indicates that the two dsDNA within the close synaptic complexes are directly ligatable. Importantly, the two ends cannot be covalently ligated within the pol μ synaptic complex in the absence of nucleotide addition (Supplementary Figure S2C). Therefore, the capability of ligation of the two duplexes can tell us the basis of the synapsis, namely whether it is pol μ-dependent or Ku plus X4L4-dependent. The results show that <25% of the substrates within the synaptic complexes formed by the combination of Ku, X4L4 and pol μ can be ligated (Figure 6E). This ligation efficiency is much less than that within the synaptic complexes formed by Ku plus X4L4 (Figure 6D). These results indicate that the majority of the synapsis events formed in the presence of Ku, X4L4 and pol μ are still pol μ-dependent.
We further tested if the pol μ BRCT domain, which plays an important role in the interaction with X4L4 (15), is required for the reversal of Ku inhibition by X4L4. The results show that X4L4 could not reverse the Ku inhibition on the pol μ ΔBRCT-mediated synapsis (Figure 6F). The results indicate that the interaction of pol μ with X4L4 is critical for the reversal of Ku inhibition by X4L4.
DISCUSSION
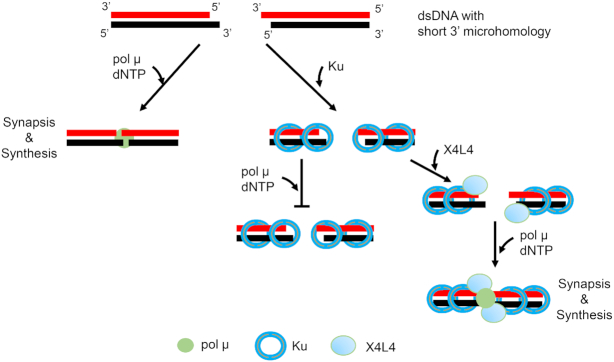
Bringing two broken dsDNA ends together is the first and most critical step for the repair of most double-strand DNA breaks and is essential for their eventual ligation. Key NHEJ proteins, Ku, X4L4 and XLF were previously shown to mediate the NHEJ synapsis of two DNA ends (6,7). Here, we directly observe that pol μ alone can mediate NHEJ synapsis of overhangs with short microhomologies. We note that pol μ was previously shown to pull-down two noncomplementary ends and carry out synthesis across the junction of noncomplementary overhangs (12,27,31). The importance of the current study is that it provides direct evidence that end microhomology is required for efficient and durable NHEJ close synapsis mediated by pol μ alone, and the synapsis is regulated by other NHEJ proteins. Based on our results, we propose an NHEJ synapsis model for pol μ (Figure 7). In cells, some broken DNA ends have 3′ overhangs with potential short microhomologies. If pol μ binds first to the DNA ends, it can mediate synapsis of the dsDNA overhangs and extend the primer in a template-dependent manner. The binding of a sub-stoichiometric Ku on the broken DNA would not affect the pol μ synapsis. But if multiple Ku proteins first occupy the entire DNA duplex, this limits the accessibility of the overhangs to pol μ, which inhibits any pol μ synapsis of that DNA end. However, if Ku is followed by X4L4 addition, then the ligase may push the end Ku protein inward, expose the overhangs and help recruit pol μ to the ends to permit pol μ to mediate NHEJ synapsis (Figure 7).
Figure 7.
NHEJ synapsis model for pol μ and the order of arrival of key factors. In cells, some of the broken DNA ends have 3′ overhangs with short microhomologies. If pol μ first binds to the DNA ends, it can mediate NHEJ synapsis of the dsDNA ends and extend the primer in a template-dependent manner. The binding of a sub-stoichiometric Ku on the broken DNA would not affect the pol μ synapsis. But if multiple Ku molecules first occupy the entire DNA duplex, this limits the accessibility of the overhangs to pol μ, which inhibits the pol μ mediated synapsis. If X4L4 then binds to Ku, it may push the Ku inward, expose the overhangs and help recruit pol μ to the ends to permit pol μ mediated synapsis. For simplicity, the NHEJ synapsis mediated by Ku, X4L4 and XLF, but without pol μ, is not shown here (6).
The direct observation of pol μ synapsis demonstrates that the synapsis step of NHEJ is as flexible as other NHEJ steps and depends on the order of arrival of NHEJ proteins at the DSB as well as the complexity of the broken ends (Supplementary Figure S8) (1,5). In a DSB, where the ends are simplest (requiring no nucleotide addition or local resection), the key NHEJ proteins, Ku, X4L4 and XLF, are sufficient to mediate NHEJ synapsis and ligation (4,6,7). If the ends are complicated by non-ligatable features, then nucleases and polymerases are required to process the ends either before NHEJ synapsis or within the NHEJ synaptic complex (Figure 7 and Supplementary Figure S8).
Pol μ NHEJ synapsis may be important, if a DSB is in a nuclear location that is locally less accessible. For example, within tightly packed heterochromatin, the molecular size of Ku70/Ku80, being over 153 kDa, may be too large to access a subset of poorly accessible DSB ends. The 56 kDa pol μ in cells may more readily diffuse to the DSB and achieve NHEJ synapsis of such broken ends. Even if Ku protein binds first to the broken ends, it can later recruit X4L4 and pol μ (14,15). Pol μ within the complex can further efficiently align the two overhangs into a physiological configuration for nucleotide addition, which can then extend terminal microhomology. The aligned and extended overhangs can then be directly ligated by X4L4 or aided by other NHEJ proteins for final ligation.
Alignment of two broken DNA ends into a proper configuration is thought to be prerequisite for pol μ to conduct template-dependent synthesis. Previous studies showed that pol μ with or without the help of other NHEJ proteins could extend the noncomplementary ends in a template-dependent manner (29,30), which suggests pol μ can mediate or participate in mediating NHEJ synapsis of these overhangs. Here, we find that at least 1 nt microhomology between the overhangs is required for pol μ alone to achieve efficient NHEJ synapsis. This is consistent with the crystal structure of the TdT-chimera NHEJ synaptic complex, in which one base-pair microhomology substrate is present (23). Only rare synaptic molecules were observed for noncomplementary overhangs. These results are consistent with a previous study showing that pol μ alone cannot efficiently extend the noncomplementary ends in a template-dependent manner and end-bridging aided by other NHEJ proteins, Ku and X4L4 is required for this extension (29). Moreover, we find that pol μ, even with the help of Ku and X4L4 cannot mediate efficient NHEJ synapsis of the incompatible overhangs used here. Other studies also have shown that pol μ alone can efficiently conduct template-dependent synthesis for some noncomplementary overhangs but not for others (19,28,30,32). In addition, our results show that a NHEJ synaptic complex formed with overhangs with 1 nt microhomology can last 0.9 s, and one additional base pairing at the ends can further stabilize and increase the lifetime of the synaptic complex by ∼2-fold. Therefore, the results here and elsewhere suggest that the capability of pol μ alone to bridge two overhangs depends on the sequence and length of the overhangs.
Pol μ has the ability to incorporate ribonucleotides at the primer end for NHEJ repair (25,26). Our results show that ribonucleotide and deoxyribonucleotide have the same capability to stimulate pol μ synapsis. The results here also demonstrate that pol μ can carry out synapsis even when the mismatched deoxyribonucleotide triphosphate is provided for the reaction (Figure 4). This capability of synapsis may explain the mis-insertion by pol μ even when the overhangs have partial microhomology (22,25).
In conclusion, our results show that pol μ by itself can mediate a durable NHEJ synapsis of overhangs with at least 1 nt microhomology, and that the initial binding of Ku and X4L4 together at the DNA ends would not affect the pol μ synapsis. The observation of pol μ synapsis provides direct evidence of diverse pathways of synapsis during NHEJ, since it can be independent of Ku:X4L4:XLF synapsis (6). Polymerase lambda (pol λ) is also an important member of the Pol X family and is more abundant than pol μ in some cells and at some stages of differentiation (5); therefore, the capability of pol λ to mediate NHEJ synapsis will be worth testing in the future. Though flexibility in the steps of NHEJ has been a theme we and others have emphasized previously (1,3,5,39,43–49), the demonstration here of multiple synaptic pathways is the clearest and first real-time demonstration of such flexibility at the NHEJ synapsis step.
DATA AVAILABILITY
All the data that support the findings of this study are available from the corresponding author upon reasonable request.
The iSMS 2.01 software was downloaded at http://inano.au.dk/about/research-groups/single-molecule-biophotonics-group-victoria-birkedal/software/.
MATLAB codes for further data analysis including EFRET and dwell distribution analysis after trajectory extraction are from Dr. Eli Rothenberg's lab at New York University School of Medicine.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Lieber lab for helpful discussions. We would like to thank Dr Marc Delarue at Unitéde Dynamique Structurale des Macromolécules, Institut Pasteur for providing helpful suggestions and proofreading an early version of the manuscript. We would also like to thank Dr Eli Rothenberg at New York University School of Medicine for sharing the codes for single molecule data analysis, and Dr Ramunas Stanciauskas from Nikon company for maintaining the TIRF microscopy. We acknowledge the Cell and Tissue Imaging Core for use of the Nikon Eclipse Ti microscope, which receives support from the USC Norris Comprehensive Cancer Center P30 grant.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [GM118009, CA196671, CA100504 to M.R.L.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Pannunzio N.R., Watanabe G., Lieber M.R.. Nonhomologous DNA end joining for repair of DNA double-strand breaks. J. Biol. Chem. 2018; 293:10512–10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scully R., Panday A., Elango R., Willis N.A.. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 2019; 20:698–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lieber M.R. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 2008; 283:1–5. [DOI] [PubMed] [Google Scholar]
- 4. Chang H.H.Y., Watanabe G., Gerodimos C.A., Ochi T., Blundell T.L., Jackson S.P., Lieber M.R.. Different DNA end configuraitons dictate which NHEJ components are most important for joining efficiency. J. Biol. Chem. 2016; 291:24377–24389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao B., Watanabe G., Morten M.J., Reid D.A., Rothenberg E., Lieber M.R.. The essential elements for the noncovalent association of two DNA ends during NHEJ synapsis. Nat. Commun. 2019; 10:3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reid D.A., Keegan S., Leo-Macias A., Watanabe G., Strande N.T., Chang H.H., Oksuz B.A., Fenyo D., Lieber M.R., Ramsden D.A. et al.. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E2575–E2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Graham T.G., Walter J.C., Loparo J.J.. Two-stage synapsis of DNA ends during non-homologous end joining. Mol. Cell. 2016; 61:850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang J.L., Duboc C., Wu Q., Ochi T., Liang S., Tsutakawa S.E., Lees-Miller S.P., Nadal M., Tainer J.A., Blundell T.L. et al.. Dissection of DNA double-strand-break repair using novel single-molecule forceps. Nat. Struct. Mol. Biol. 2018; 25:482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moon A.F., Garcia-Diaz M., Batra V.K., Beard W.A., Bebenek K., Kunkel T.A., Wilson S.H., Pedersen L.C.. The X family portrait: structural insights into biological functions of X family polymerases. DNA Repair (Amst.). 2007; 6:1709–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Domínguez O., Ruiz J.F., Laín de Lera T., García-Díaz M., González M.A., Kirchhoff T., Martínez-A C., Bernad A., Blanco L.. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000; 19:1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nick McElhinny S.A., Havener J.M., Garcia-Diaz M., Juárez R., Bebenek K., Kee B.L., Blanco L., Kunkel T.A., Ramsden D.A.. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol. Cell. 2005; 19:357–366. [DOI] [PubMed] [Google Scholar]
- 13. Gu J., Lu H., Tippin B., Shimazaki N., Goodman M.F., Lieber M.R.. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007; 26:1010–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mahajan K.N., Nick McElhinny S.A., Mitchell B.S., Ramsden D.A.. Association of DNA polymerase μ (pol μ) with Ku and ligase IV: role for pol μ in end-joining double-strand break repair. Mol. Cell Biol. 2002; 22:5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma Y., Lu H., Tippin B., Goodman M.F., Shimazaki N., Koiwai O., Hsieh C.-L., Schwarz K., Lieber M.R.. A biochemically defined system for mammalian nonhomologous DNA end joining. Mol. Cell. 2004; 16:701–713. [DOI] [PubMed] [Google Scholar]
- 16. Bertocci B., De Smet A., Berek C., Weill J.-C., Reynaud C.-A.. Immunoglobulin κ Light chain gene rearrangement is impaired in mice deficient for DNA polymerase Mu. Immunity. 2003; 19:203–211. [DOI] [PubMed] [Google Scholar]
- 17. Lieber M.R. The polymerases for V(D)J Recombination. Immunity. 2006; 25:7–9. [DOI] [PubMed] [Google Scholar]
- 18. Loc’h J., Delarue M.. Terminal deoxynucleotidyltransferase: the story of an untemplated DNA polymerase capable of DNA bridging and templated synthesis across strands. Curr. Opin. Struct. Biol. 2018; 53:22–31. [DOI] [PubMed] [Google Scholar]
- 19. Martin M.J., Juarez R., Blanco L.. DNA-binding determinants promoting NHEJ by human Polμ. Nucleic Acids Res. 2012; 40:11389–11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nick McElhinny S.A., Ramsden D.A.. Polymerase Mu is a DNA-directed DNA/RNA polymerase. Mol. Cell Biol. 2003; 23:2309–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moon A.F., Pryor J.M., Ramsden D.A., Kunkel T.A., Bebenek K., Pedersen L.C.. Sustained active site rigidity during synthesis by human DNA polymerase μ. Nat. Struct. Mol. Biol. 2014; 21:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moon A.F., Garcia-Diaz M., Bebenek K., Davis B.J., Zhong X., Ramsden D.A., Kunkel T.A., Pedersen L.C.. Structural insight into the substrate specificity of DNA polymerase μ. Nat. Struct. Mol. Biol. 2007; 14:45–53. [DOI] [PubMed] [Google Scholar]
- 23. Loc’h J., Gerodimos C.A., Rosario S., Tekpinar M., Lieber M.R., Delarue M.. Structural evidence for an in trans base selection mechanism involving Loop1 in polymerase μ at an NHEJ double-strand break junction. J. Biol. Chem. 2019; 294:10579–10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ruiz J.F., Juárez R., García‐Díaz M., Terrados G., Picher A.J., González‐Barrera S., Fernández de Henestrosa A.R., Blanco L.. Lack of sugar discrimination by human Pol μ requires a single glycine residue. Nucleic Acids Res. 2003; 31:4441–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin M.J., Garcia-Ortiz M.V., Esteban V., Blanco L.. Ribonucleotides and manganese ions improve non-homologous end joining by human Polμ. Nucleic Acids Res. 2012; 41:2428–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pryor J.M., Conlin M.P., Carvajal-Garcia J., Luedeman M.E., Luthman A.J., Small G.W., Ramsden D.A.. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science. 2018; 361:1126–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Juárez R., Ruiz J.F., McElhinny S.A.N., Ramsden D., Blanco L.. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006; 34:4572–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martin M.J., Blanco L.. Decision-making during NHEJ: a network of interactions in human Polμ implicated in substrate recognition and end-bridging. Nucleic Acids Res. 2014; 42:7923–7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davis B.J., Havener J.M., Ramsden D.A.. End-bridging is required for pol μ to efficiently promote repair of noncomplementary ends by nonhomologous end joining. Nucleic Acids Res. 2008; 36:3085–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Andrade P., Martín M.J., Juárez R., López de Saro F., Blanco L.. Limited terminal transferase in human DNA polymerase μ defines the required balance between accuracy and efficiency in NHEJ. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:16203–16208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gouge J., Rosario S., Romain F., Poitevin F., Béguin P., Delarue M.. Structural basis for a novel mechanism of DNA bridging and alignment in eukaryotic DSB DNA repair. EMBO J. 2015; 34:1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin M.J., Garcia-Ortiz M.V., Gomez-Bedoya A., Esteban V., Guerra S., Blanco L.. A specific N-terminal extension of the 8 kDa domain is required for DNA end-bridging by human Polμ and Polλ. Nucleic Acids Res. 2013; 41:9105–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Loc’h J., Rosario S., Delarue M.. Structural basis for a new templated activity by terminal deoxynucleotidyl Transferase: Implications for V(D)J recombination. Structure. 2016; 24:1452–1463. [DOI] [PubMed] [Google Scholar]
- 34. Lee W., von Hippel P.H., Marcus A.H.. Internally labeled Cy3/Cy5 DNA constructs show greatly enhanced photo-stability in single-molecule FRET experiments. Nucleic Acids Res. 2014; 42:5967–5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Preus S., Noer S.L., Hildebrandt L.L., Gudnason D., Birkedal V.. iSMS: single-molecule FRET microscopy software. Nat. Methods. 2015; 12:593–594. [DOI] [PubMed] [Google Scholar]
- 36. Preus S., Hildebrandt L.L., Birkedal V.. Optimal background estimators in single-molecule FRET microscopy. Biophys. J. 2016; 111:1278–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zagelbaum J., Shimazaki N., Esguerra Z.A., Watanabe G., Lieber M.R., Rothenberg E.. Real-time analysis of RAG complex activity in V(D)J recombination. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11853–11858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moon A.F., Gosavi R.A., Kunkel T.A., Pedersen L.C., Bebenek K.. Creative template-dependent synthesis by human polymerase mu. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4530–E4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pryor J.M., Waters C.A., Aza A., Asagoshi K., Strom C., Mieczkowski P.A., Blanco L., Ramsden D.A.. Essential role for polymerase specialization in cellular nonhomologous end joining. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:E4537–E4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reid D.A., Conlin M.P., Yin Y., Chang H.H., Watanabe G., Lieber M.R., Ramsden D.A., Rothenberg E.. Bridging of double-stranded breaks by the nonhomologous end-joining ligation complex is modulated by DNA end chemistry. Nucleic Acids Res. 2017; 45:1872–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. de Vries E., van Driel W., Bergsma W.G., Arnberg A.C., van der Vliet P.C.. HeLa nuclear protein recognizing DNA termini and translocating on DNA forming a regular DNA-multimeric protein complex. J. Mol. Biol. 1989; 208:65–78. [DOI] [PubMed] [Google Scholar]
- 42. Kysela B., Doherty A.J., Chovanec M., Stiff T., Ameer-Beg S.M., Vojnovic B., Girard P.M., Jeggo P.A.. Ku stimulation of DNA ligase IV-dependent ligation requires inward movement along the DNA molecule. J. Biol. Chem. 2003; 278:22466–22474. [DOI] [PubMed] [Google Scholar]
- 43. Conlin M.P., Reid D.A., Small G.W., Chang H.H., Watanabe G., Lieber M.R., Ramsden D.A., Rothenberg E.. DNA ligase IV guides End-Processing choice during nonhomologous End joining. Cell Rep. 2017; 20:2810–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lieber M.R., Gu J., Lu H., Shimazaki N., Tsai A.G.. Nonhomologous DNA end joining (NHEJ) and chromosomal translocations in humans. Subcell. Biochem. 2010; 50:279–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gu J., Lieber M.R.. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008; 22:411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lieber M.R., Lu H., Gu J., Schwarz K.. Flexibility in the order of action and in the enzymology of the nuclease, polymerases, and ligase of vertebrate non-homologous DNA end joining: relevance to cancer, aging, and the immune system. Cell Res. 2008; 18:125–133. [DOI] [PubMed] [Google Scholar]
- 47. Gu J., Lu H., Tsai A.G., Schwarz K., Lieber M.R.. Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence. Nucleic Acids Res. 2007; 35:5755–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ma Y., Lu H., Schwarz K., Lieber M.R.. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005; 4:1193–2000. [DOI] [PubMed] [Google Scholar]
- 49. Ma Y., Schwarz K., Lieber M.R.. The artemis:DNA-PKcs endonuclease can cleave gaps, Flaps, and Loops. DNA Repair (Amst.). 2005; 4:845–851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data that support the findings of this study are available from the corresponding author upon reasonable request.
The iSMS 2.01 software was downloaded at http://inano.au.dk/about/research-groups/single-molecule-biophotonics-group-victoria-birkedal/software/.
MATLAB codes for further data analysis including EFRET and dwell distribution analysis after trajectory extraction are from Dr. Eli Rothenberg's lab at New York University School of Medicine.