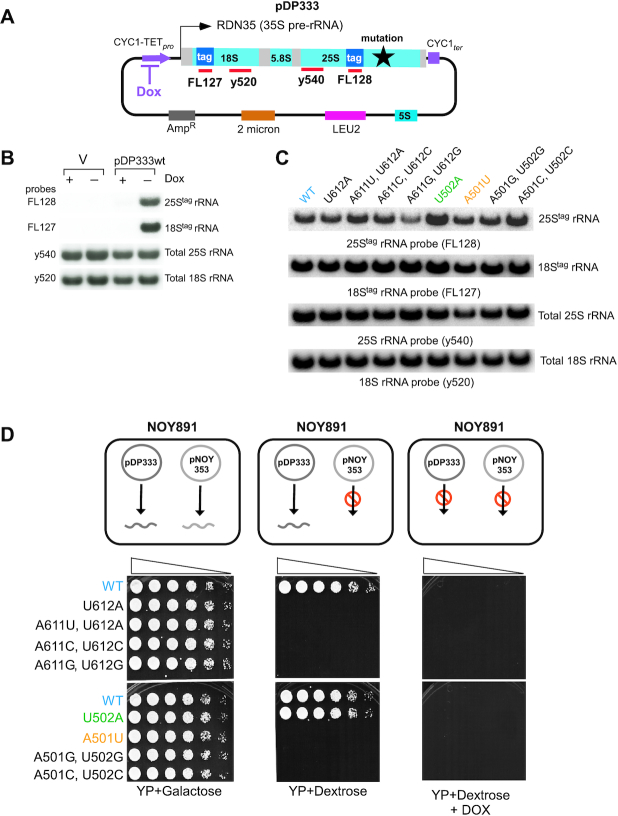
Figure 2.
Design, validation of expression, and viability of mutant tagged rRNAs. (A) Schematic representation of the pDP333 plasmid. Doxycycline (Dox)-repressible promoter (CYC1-TETpro) and CYC1 terminator (CYC1ter) flank the 35S pre-rRNA coding sequence RDN35. Probes FL127 and FL128 allow specific detection of plasmid-derived rRNAs through short tag sequences inserted into the 18S and 25S rRNA coding sequences, respectively. Probes y520 and y540 detect both endogenous and plasmid-derived rRNA. pDP333 also carries a copy of the RDN5 gene for 5S rRNA, a LEU2 selectable marker, 2μ DNA replication origin and ampicillin resistance gene. (B) BY4741 cells transformed with pNS1 (empty vector control, V) or pDP333 containing tagged wild-type rRNA were grown in SD leucine dropout (SD-leu-) media with (+) or without (−) Dox, total RNA was extracted and analyzed by northern hybridizations with probes indicated on the left. (C) BY4741 cells transformed with pDP333 plasmids containing the indicated mutations were grown as in (B) without Dox, RNA was analyzed by northern hybridizations. (D) Top panel: Using media of different composition with the NOY891 strain allows two different plasmids, pDP333 and pNOY353, to be used as the source of rRNA. Bottom panel: NOY891 strain carrying pNOY353 to express wild-type rRNAs from a GAL1 promoter was transformed with pDP333 plasmids carrying mutations indicated on the left. Transformants were grown in SD-leu-ura- in the presence of Dox at 30°C, adjusted to the same cell density and their five-fold dilutions were plated on agar media indicated at the bottom. Plates were incubated at 30°C for 5–7 days.