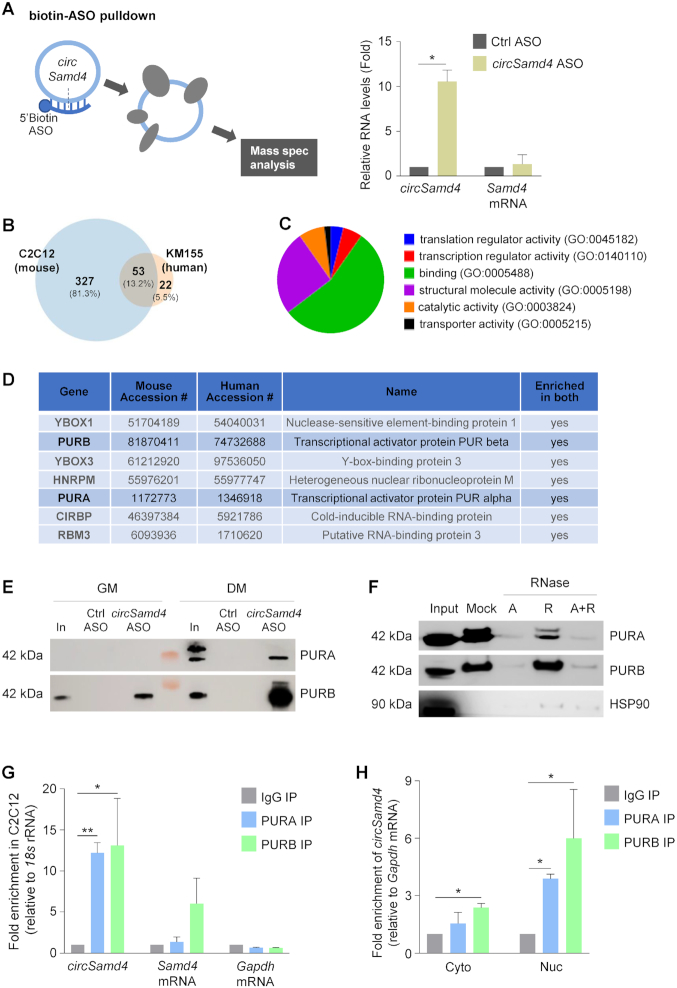
Figure 4.
circSamd4 interacts with transcription factors PURA and PURB. (A) Left, biotinylated antisense oligomers (ASOs) complementary to the junction of circSamd4 and control (Ctrl) were incubated with C2C12 in GM as well as in DM. After affinity pulldown using streptavidin beads, circSamd4-interacting proteins were identified by mass spectroscopy. Right, levels of circSamd4 enrichment in ASO pulldown samples, as assessed by RT-qPCR analysis of circSamd4 (relative to the enrichment of Gapdh mRNA, a transcript that does not bind the ASOs and encodes a housekeeping protein) in the pulldown samples. Enrichment in Samd4 mRNA was monitored in parallel. (B) Venn diagram representation of the proteins significantly enriched in the mass spec datasets identified after pulldown of circSamd4 ASO vs Ctrl ASOs in mouse C2C12 myoblasts (left) and in human KM155 myoblasts (right) (Supplementary Figure S3; Materials and Methods). 53 proteins were shared in both mass spec datasets (intersection). (C) Pie chart represents GO functional categories of the 53 proteins at the intersection of both datasets. (D) Top 7 proteins shared in the mass spec datasets from mouse and human myoblast pulldowns (Supplementary Table S2 and Supplementary Figure S3A and S3B). (E) Western blot analysis of the interaction of circSamd4 with PURA and PURB present in DM and GM lysates, as assessed after biotin-ASO pulldown followed by western blot analysis of PURA and PURB levels in the pulldown material. Input (In), 5 μg of GM or DM lysates (C2C12). (F) The interaction of PURA and PURB with circSamd4 was assessed as explained in panel (E), except that lysates were treated with RNase A and/or RNase R before pulldown followed by western blot analysis. (G) For RIP (ribonucleoprotein immunoprecipitation) analysis, IP was performed using DM C2C12 lysates and either IgG (control) antibodies or antibodies recognizing PURA or PURB; the presence of circSamd4 and Samd4 mRNA (as well as control Gapdh mRNA) in the IP material was measured by RT-qPCR analysis. Enrichments were normalized to the levels of 18s rRNA. (H) After fractionating nuclei and cytoplasm, RIP analysis was carried out as explained in (G) to measure the interaction of PURA and PURB with circSamd4 in the nucleus and the cytoplasm. Data in (A,G,H) are the means ±SEM from three independent experiments. *P < 0.05, **P < 0.005.