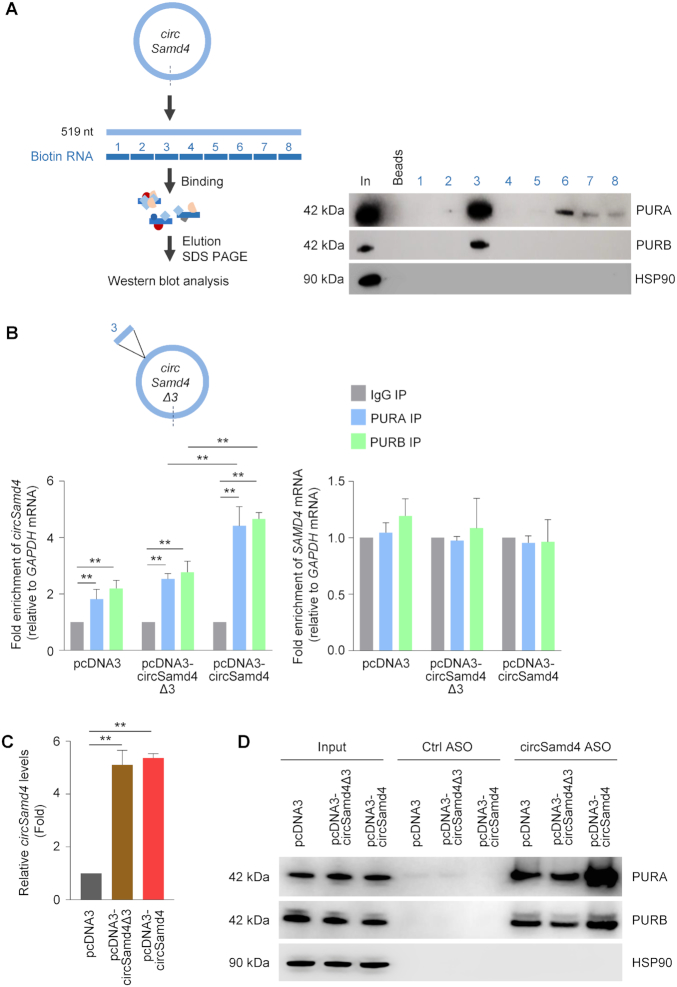
Figure 5.
PUR proteins predominantly associate with a specific region of circSamd4. (A) Schematic of biotin pulldown of PURA and PURB using nonoverlapping biotinylated RNAs (each ∼65 nt long) tiling the length of circSamd4. Following incubation of the biotinylated RNAs with C2C12 DM lysates, bound proteins were pulled down using streptavidin beads and the presence of PURA and PURB was assessed by western blot analysis. Input (In) was 2 μg of DM lysates; Beads only (Beads), no biotinylated RNA. (B, C) Schematic, circSamd4Δ3 bearing a deletion of segment 3. By 24 h after transfecting HeLa cells with vectors (2 μg) to overexpress circSamd4 and circSamd4(Δ3), RIP analysis was used to measure circSamd4 (left graph) and SAMD4 mRNA (right graph) bound to PURA and PURB relative to the levels of GAPDH mRNA (B). The levels of circSamd4Δ3 and circSamd4 expressed in HeLa cells 24 h after transfection of plasmids pcDNA3-circSamd4Δ3 and pcDNA3-circSamd4, respectively, were measured by RT-qPCR analysis and normalized to GAPDH mRNA levels (C). (D) Following overexpression of circSamd4 and circSamd4Δ3 as explained in panel (B), biotin-ASO pulldown was carried out to confirm the interaction of PURA and PURB with circSamd4 in C2C12 lysates as shown by Western blot analysis. The levels of the housekeeping protein HSP90 were assessed as a background control protein. Input was 2 μg of DM lysates. Data in (B,C) are the means ±SEM from three independent experiments. **P < 0.005.