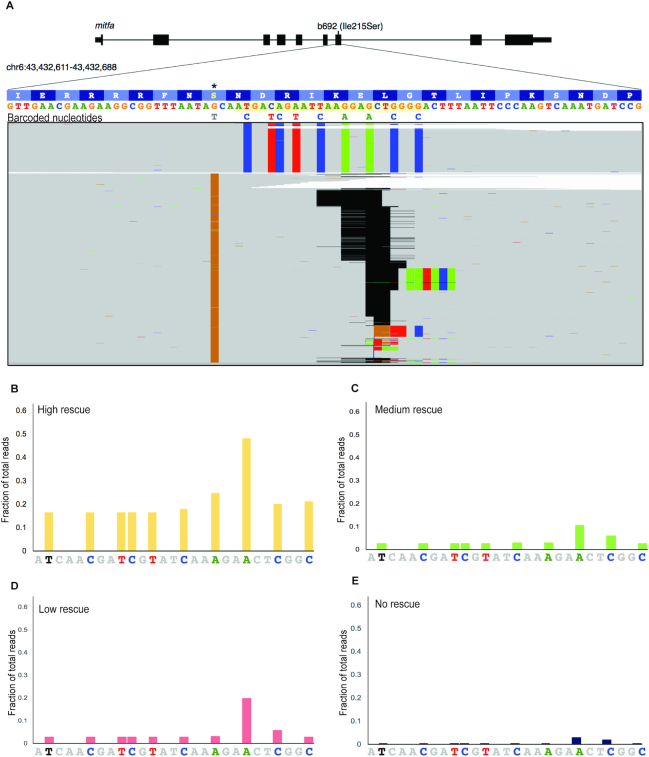
Figure 3.
Quantitative assessment of genome editing efficiency by next-generation sequencing. (A) Alignment of next generation sequencing reads from b692 genome edited embryos from the high rescue phenotype. Exon 7 of mitfa is displayed. The gRNA sequence is highlighted in yellow. b692 mutants have a T>G mutation leading to an isoleucine to serine substitution at codon 215(*). The 951bp HDR template restores the wild-type isoleucine codon (ATC) and encodes nine additional barcoded nucleotides as indicated. Indels in sequencing reads are represented as gapped regions in black. (B–E) The fraction of reads at each barcoded nucleotide in the HDR template is plotted for each phenotype category.