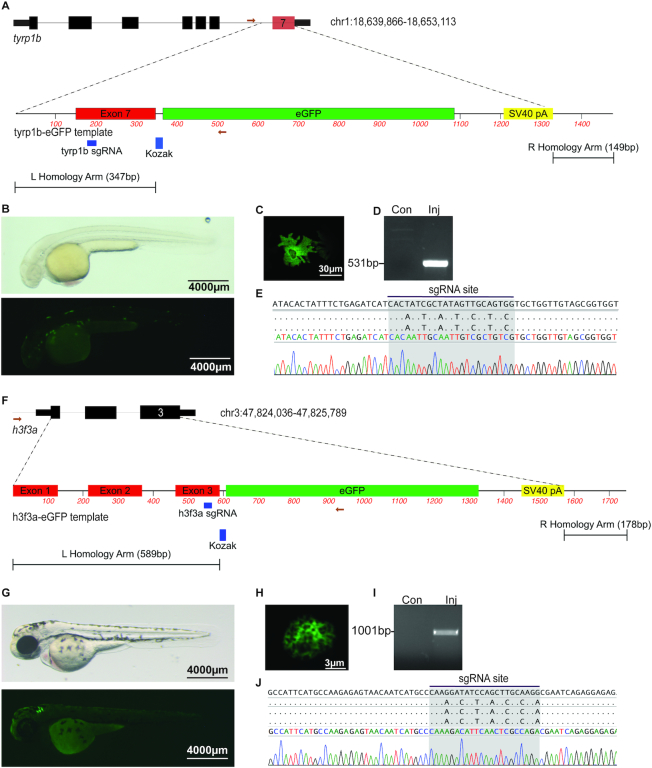
Figure 4.
Genome editing leads to precise fluorophore knock-in. (A) Schematic depicting tyrp1b locus and tyrp1b-EGFP template. The gRNA recognition sequence in exon 7 is noted. (B) albino(b4) embryo injected with rCas9, tyrp1b synthetic gRNA, and the tyrp1b-EGFP template dsDNA template, light (top panel) or fluorescence (bottom panel) microscopy at 48hpf. (C) Confocal imaging reveals the presence of stellate EGFP-positive melanocyte. (D) Allele-specific PCR was performed on uninjected or injected/GFP+ embryos at 48hpf. (E) Sanger sequencing from allele-specific PCR detects template integration and modified nucleotides at the gRNA recognition site. (F) Schematic depicting h3f3a locus and h3f3a-eGFP template. The gRNA recognition sequence in exon 3 is noted. (G) Wild-type embryo injected with rCas9, h3f3a gRNA, and dsDNA template is photographed with light (top panel) or fluorescence (bottom panel) microscopy (48hpf). (H) Confocal imaging reveals the presence of an EGFP-positive nuclei. (I) Allele-specific PCR was performed on uninjected or HDR injected/GFP+ embryos at 48hpf. (J) Sanger sequencing was performed from allele-specific PCR to detect template integration. Barcoded nucleotides at the gRNA binding site are identified.