Figure 2.
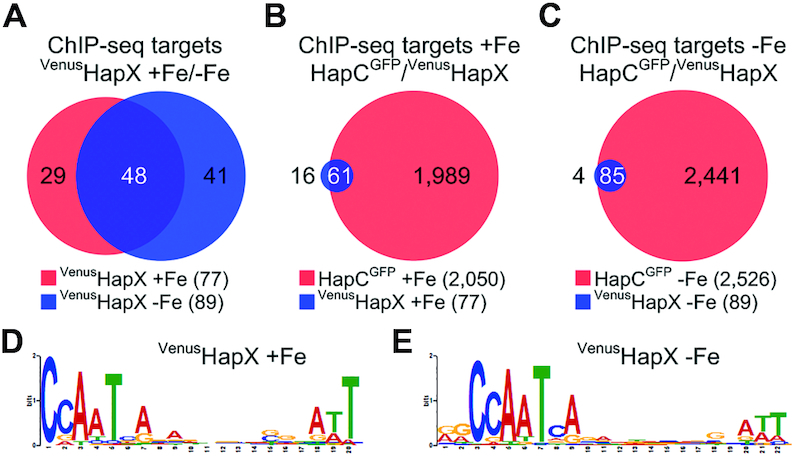
Identification of common direct targets of HapX and the CBC by genome-wide ChIP-seq analysis and de novo discovery of a CBC:HapX motif. Venn diagrams showing comparison of ChIP-seq target sites of HapX and the CBC were depicted using a modified version of Cistrome software (47). For comparison of common ChIP-seq target sites, 150 bp sequences centered at the peak summit regions were used. The total number of ChIP-seq target sites is reported in parentheses. Numbers in white indicate how many ChIP sequence targets overlap in the respective comparison. (A) Comparison of the VenusHapX ChIP-seq target sites (>3-fold enrichment, found within the 1.5-kb of the 5′-upstream region) during iron sufficiency (+Fe) versus iron starvation (−Fe) conditions. (B) Comparison of the HapCGFP and the VenusHapX ChIP-seq target sites (>2-fold enrichment for the CBC, >3-fold enrichment for HapX, found within 1.5 kb of the 5′-upstream region) during +Fe conditions. (C) Comparison of the HapCGFP and the VenusHapX ChIP-seq target sites during −Fe conditions. CBC:HapX consensus DNA recognition motifs identified by MEME from the VenusHapX ChIP-seq peak summit regions for +Fe (D) and −Fe (E) conditions.
