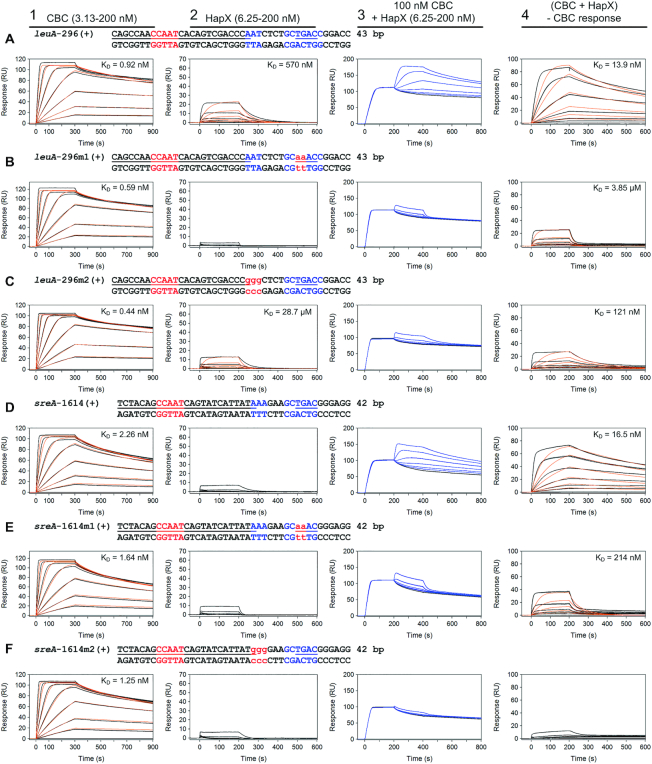
Figure 6.
HapX-binding depends in vitro on both the Gcn4-like half sites and A/T-rich submotifs within the bipartite CBC:HapX consensus sites as shown by SPR analysis. SPR analyses included binding of the CBC to DNA (panel 1), HapX to DNA (panel 2) and HapX to preformed CBC:DNA complexes (panel 3). The SPR sensorgrams are shown from sensor-immobilized duplexes covering evolutionary conserved bipartite CBC:HapX binding sites from A. fumigatus leuA and sreA promoters (A and D) as well as duplexes carrying mutations within their 3′ Gcn4-like half-site (B and E) or A/T-rich submotif (C and F). Nt underlined in black are covered by the CBC and nt marked in blue represent the A. fumigatus conserved submotifs. Gcn4-like half-sites are underlined in blue. Substituted nt relative to the wild type sequence are written in red and shown in lower case. Binding responses of the indicated CBC or HapX concentrations injected in duplicate (black lines) are shown overlaid with the best fit derived from a 1:1 interaction model including a mass transport term (red lines). Binding responses of CBC:DNA:HapX ternary complex formation (panel 3, blue lines) were obtained by concentration dependent co-injection of HapX on preformed binary CBC:DNA complexes after 200 seconds within the steady-state phase. Sensorgrams in panel 4 depict the association/dissociation responses of HapX on preformed CBC:DNA and were generated by CBC response subtraction (co-injection of buffer) from HapX co-injection responses.