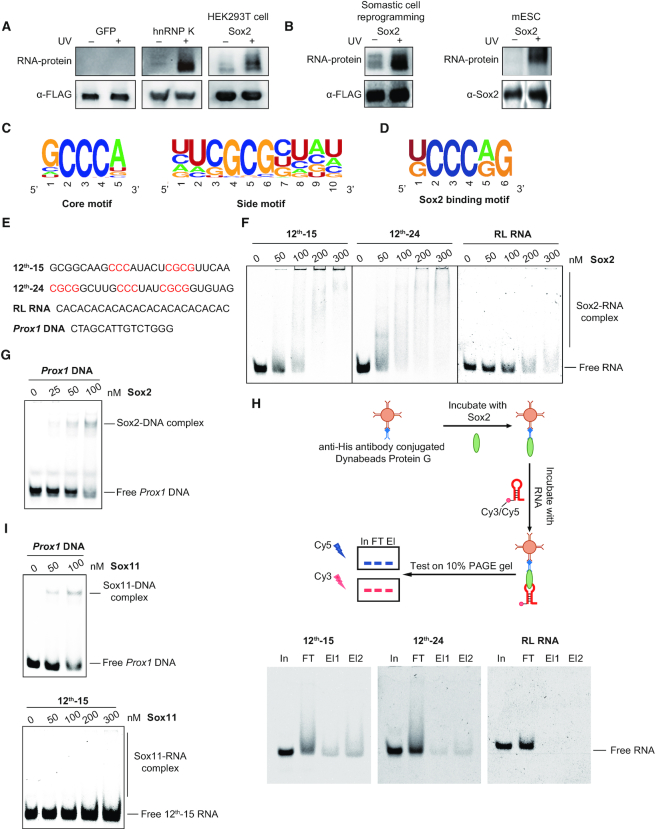
Figure 1.
Sox2 preferentially binds G/C-rich RNA sequences. (A) RNA binding activity of Sox2 was detected by PAR-CLIP. The upper panel shows the protein-bound RNA detected by biotin chemiluminescence. Antibody against FLAG tag was used to confirm the protein size and as loading control. FLAG-tagged Sox2, GFP (negative control) and hnRNP K (positive control) were expressed in HEK293T cells. (B) Same as in (A), FLAG-tagged Sox2 was expressed in MEFs at day 6 of pluripotency reprogramming, and endogenous Sox2 antibody was used in mESCs. Antibody against Sox2 or FLAG tag was used to confirm the protein size and as loading control. (C) Consensus motifs discovered by MEME in sequences enriched after 12 rounds of RNA SELEX with full length His6-Sox2 as bait (Supplementary Table S2). (D) Consensus motifs found by MEME in PAR-CLIP sequencing data for Sox2 expressed in HEK293T cells. (E) Nucleic acid ligands used for the EMSAs in (F–I). Bases highlighted in red are the SELEX consensus. 12th-15 RNA and 12th-24 RNA are Cy3- or Cy5-labelled RNA selected by SELEX after 12-round enrichment; RL RNA is a randomly chosen CA-rich RNA from the original RNA library; Prox1 DNA is a Cy5-labelled DNA with Sox2-binding motif. (F) EMSAs for the binding of 0–300 nM Sox2 to 50 nM fluorescently-labelled RNAs. The concentrations of Sox2 are as indicated above each panel. Shift patterns of free RNA and RNA bound by Sox2 (Sox2-RNA complex) are labelled to the right. (G) EMSAs of Cy5-labelled Prox1 DNA (50 nM) with increasing concentrations (0–100 nM) of Sox2. (H) In vitro RIP with 100 nM Sox2 and 100 nM fluorescently-labelled RNAs. In: input; FT: flow-through; El: elution. (I) EMSAs of Cy5-labelled Prox1 DNA (50 nM) (left panel) or Cy3-labelled 12th-15 RNA (50 nM) (right panel) with increasing concentrations of Sox11.