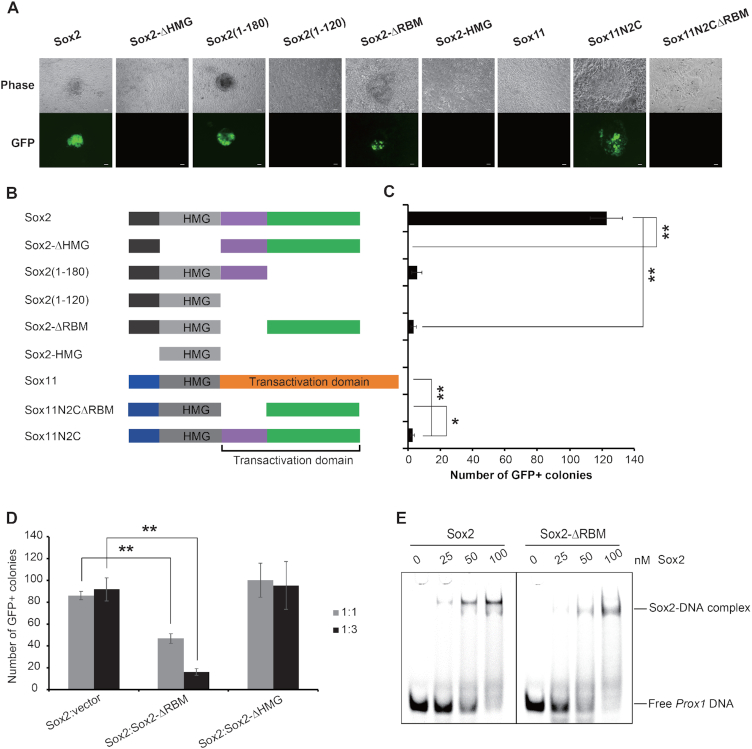
Figure 5.
Deletion of the Sox2 RBM impairs iPSC generation. (A) Morphology of mouse iPSCs generated by Sox2 or Sox11 constructs in combination with Oct4, c-Myc and Klf4 under LIF/Serum conditions generated from OG2 MEFs expressing a transgenic Oct4-GFP reporter. Colonies were photographed using a fluorescence microscope 12 days after the viral infection. The scale bar indicates 100 μm. (B) A schematic representation of Sox2 and Sox11 constructs used for iPSCs generation. (C) The iPSC generation efficiency of these constructs is presented by the absolute number of GFP-positive colonies in a well of a 12-well cell culture. Barplot shows the mean ± SD of three biological replicates and differences between selected samples were compared using ANOVA (* and ** indicate P-values of < 0.05 and < 0.01, respectively). (D) The numbers of iPSC colonies generated by Sox2 plus empty vector, Sox2-ΔRBM or Sox2-ΔHMG at 1:1 or 1:3 ratio of viral supernatants from three independent experiments are shown. Data are represented as mean ± SD, and ANOVA was used to assess significance (** indicates P-value of < 0.01). (E) Comparison of the binding activities of Sox2 and Sox2-ΔRBM to Prox1 DNA. The reactions were analyzed by EMSAs in 10% native PAGE gels. Concentrations of Sox2 constructs are labelled above each lane. Free DNA and protein-DNA complex are marked.