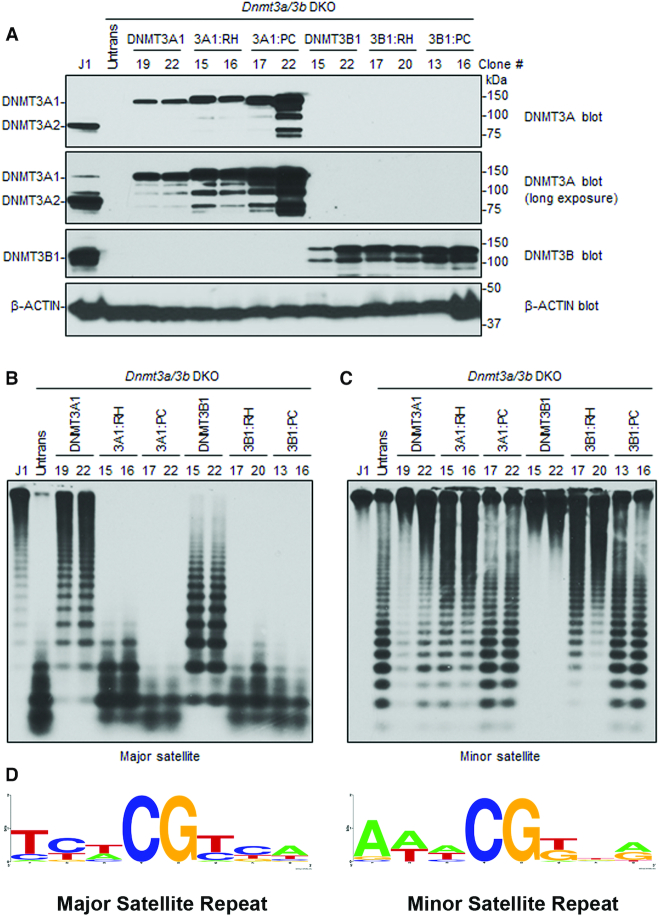
Figure 7.
Rescue of DNA methylation at the major and minor satellite repeats in Dnmt3a/3b DKO mESCs. (A) Dnmt3a/3b DKO mESCs were transfected with plasmids encoding mouse DNMT3A1 WT, DNMT3A1 Arg878His (3A1:RH), DNMT3A Pro705Val/Cys706Asp (3A1:PC), DNMT3B1 WT, DNMT3B1 Arg829His (3B1:RH), or DNMT3B1 Pro656Gly/Cys657Thr (3B1:PC), and stable clones were derived. Total cell lysates were used to analyze the expression of DNMT3A or DNMT3B proteins by western blot with anti-DNMT3A, anti-DNMT3B, and anti-β-Actin antibodies. A long exposure of the DNMT3A blot is included to show endogenous DNMT3A1 in WT (J1) mESCs. Note that stable clones showing similar expression levels to those of endogenous DNMT3A or DNMT3B were used for the experiments. (B, C) DNA methylation was analyzed by Southern blot. Genomic DNA was digested with MaeII (major satellite repeats) or HpaII (minor satellite repeats), and probed for the major (B) or minor (C) satellite repeats. J1 (WT) and untransfected DKO mESCs were used as controls. The numbers on the top indicate clone#. Complete digestion due to low or no DNA methylation is indicated by low molecular weight bands as seen in untransfected DKOs, and high molecular weight bands as seen in J1 indicate high DNA methylation and protection from digestion. Comparing the activity of DNMT3A1 clones 19/22 with 3A1:RH clones 15/16 at major and minor satellite repeats shows that 15/16 methylate minor repeats similar to 19/22 whereas at major satellite repeats the activity of 15/16 is severely impaired. (D) Consensus sequence of the nucleotides flanking the CpG site in either the major or minor satellite repeats, created using WebLogo shows high prevalence G at N+1 and N+3 positions at minor repeats.