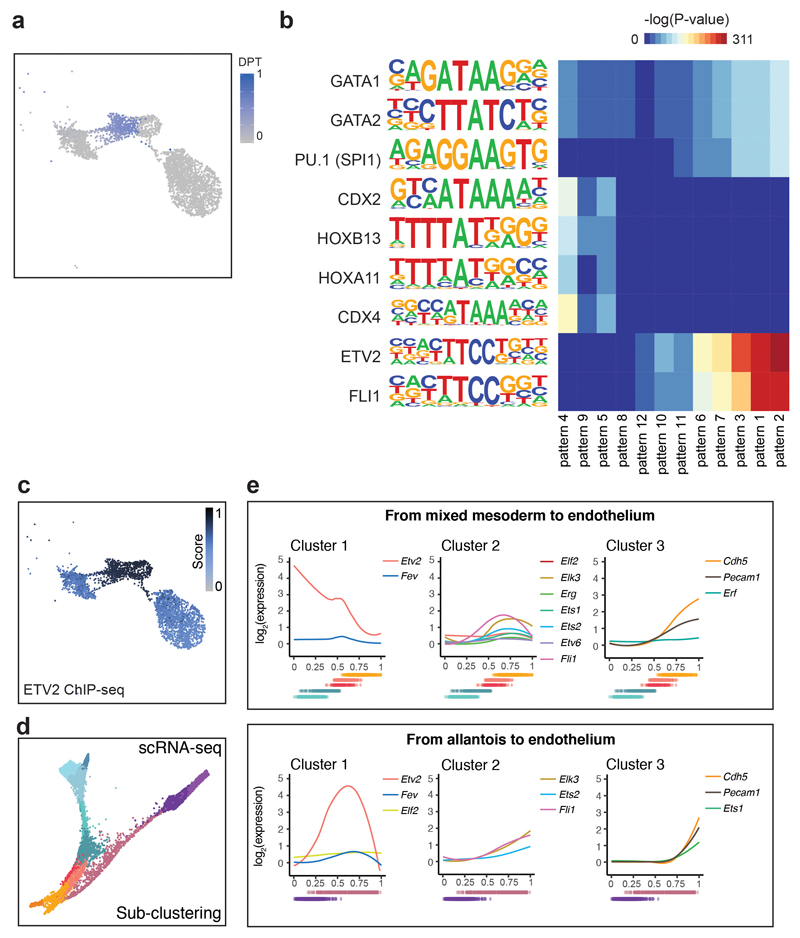
Extended Data Fig. 9. Endothelial development from allantoic cells.
a, UMAP visualisation (n=3,284 cells) showing the pseudotime trajectory from allantois to endothelium as a gradient from grey to blue. Cells scored as 0 in the plot (grey) were not part of the trajectory. b, Heatmap showing the -log(P-value) obtained from a TF motif enrichment analysis on the accessibility patterns found in Fig. 7b using HOMER. -log(P-value) ranges from 0 (dark blue) to 311 (dark red). c, UMAP visualisation of the allantoic-haemato-endothelial landscape (n=3,284 cells) showing the enrichment score (from grey=0 to dark blue=1) for ETV2 ChIP-seq peaks from41. d, Force-directed graph showing cells from the “Mixed mesoderm”, “Allantois”, “Haemato-endothelial progenitors” and “Endothelium” that have been profiled with scRNA-seq in8 (n=7,631 cells). Cell colours show the different sub-clusters found when re-analysing this dataset. e, Expression dynamics of highly variable ETS factors (variance > 0.15) along the trajectory from mixed mesoderm to endothelium (top) and from allantois to endothelium (bottom). Cdh5 and Pecam1 have been added as positive controls for mature endothelium. Dots below plots represent the ordered cells coloured by the sub-clusters in panel (d).