Figure 9.
GCN2-Induced Regulation of the mRNA Translation State.
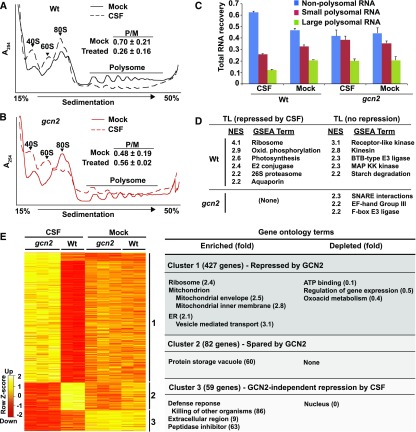
Cell extracts from Arabidopsis seedling shoots treated with 0.5 µM chlorosulfuron (2 h) were fractionated over Suc gradients to separate polysomal from nonpolysomal RNAs.
(A) and (B) UV light absorbance profiles of polysome gradients. Upon activation by CSF, GCN2 kinase inhibits polysome loading in the wild type but not in gcn2. The ratio of polysomes (P) to monosomes (M) is indicated with SE from three replicates.
(C) Gradient fractions were pooled into nonpolysomal (NP), small (SP), and large polysomal (LP) RNA pools, respectively. The histogram shows the average RNA recovered with SE.
(D) Gene set enrichment analysis of mRNA translation state. The differential translations states of 13,551 mRNAs were rank-ordered for each of two pairwise comparisons (wild type ± CSF, gcn2 ± CSF). Next, 257 gene sets harboring between 15 and 500 members were examined for a biased distribution along the rank-ordered mRNAs. Gene sets with a bias passing a family-wise error rate < 0.05 are listed with their normalized enrichment score (NES) where 0 equals no enrichment.
(E) The NP, SP, and LP RNA samples were processed for microarray gene expression profiling. Limma with FDR correction (P < 0.05) identified 568 differentially translated genes. Translation state values are displayed as a Z score. Three major clusters were identified and analyzed for functional enrichment or depletion using topGO (for details, see Supplemental Table 1).