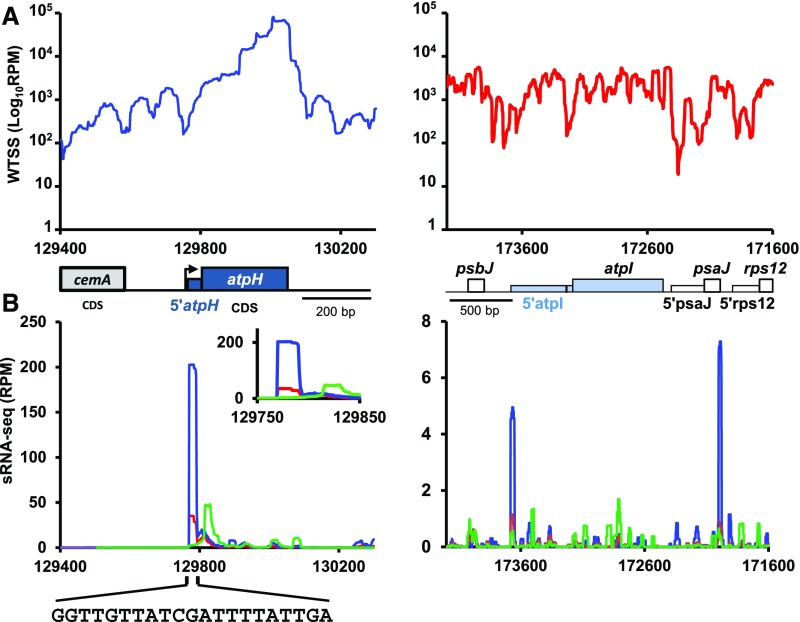
Figure 10.
Transcriptional Profiles of the atpH and atpI Genes.
(A) Coverage, normalized as RPM (log scale) of pooled the bidirectional and directional wild-type whole transcriptome shotgun sequencing (WTSS) along the atpH (left) and atpI (right) loci. Positions of the relevant genes and 5′ UTR are shown below. The black bar in the atpI 5′ UTR shows the position of the MTHI1 target (see below). Redrawn from the data in Cavaiuolo et al. (2017).
(B) sRNA mapping to the 5′ end of atpH mRNA is the footprint of MTHI1. Coverage, normalized as RPM, of pooled sRNA-seq along the same loci: the mock- (green) versus RPP-treated (blue) wild-type sRNA-seq libraries compared with RPP-treated libraries of the mthi1-1 mutant (red). Coverage is averaged over two biological replicates (libraries prepared from two independent RNA samples). Only reads mapping to the coding strand are shown. The inset for atpH shows a close-up view of the atpH 5′ UTR, and the sequence of the atpH footprint is shown. For atpI, a close-up view of the 5′ UTR (coding strand only) is shown in Supplemental Figure 9A. Note the very different values on y axes of the two graphs.