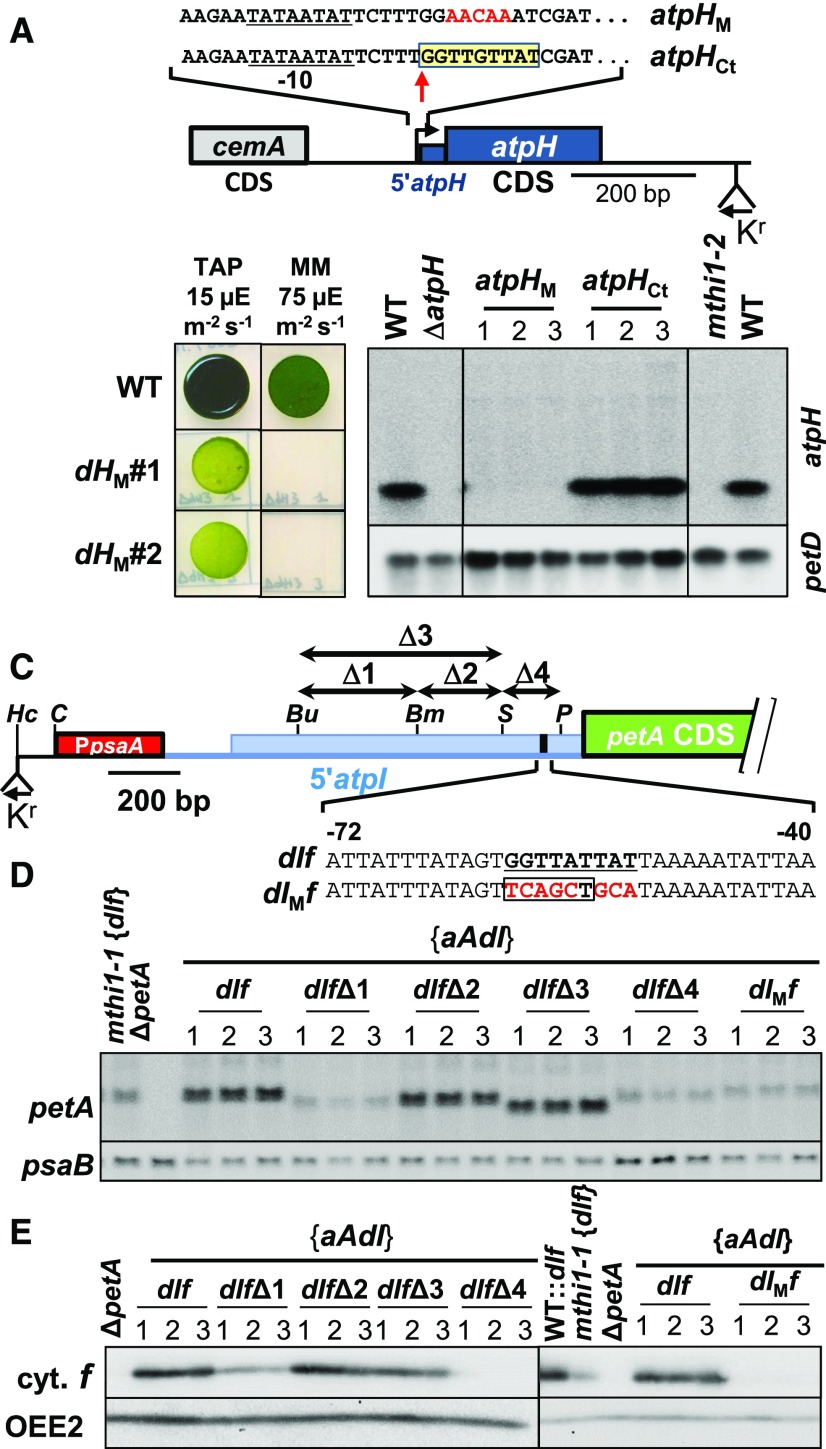
Figure 13.
Validation of the Putative MTHI1 Targets.
(A) Schematic map of the atpHM construct with a close-up view of the region of the MTHI1 binding site, highlighted in a yellow box. Mutated nucleotides are written in red. The atpH transcription start site is indicated by a vertical arrow. The atpH promoter is underlined, and the position of the recycling aadA cassette is shown. The control construct (atpHCt) carries the selection cassette but no mutation in the atpH gene.
(B) (Left) Phototrophic growth of the atpHM strain (two independent transformants), assessed as in Figure 2B. Growth of the wild type (WT) is shown as a control. MM, minimal medium. (Right) Accumulation of the atpH transcript in the wild type transformed by the atpHCt and atpHM constructs, assessed by RNA gel blots. Three independent transformants are shown for each genetic background. The petD transcript is provided as a loading control.
(C) Schematic representation of the 5′atpI 5′ UTR in the mutant dIf series.
The red rectangle represents the psaA promoter region, and the blue line shows the psbJ-atpI intergenic fragment inserted in the construct (larger than the atpI 5′ UTR, to allow the processing of the chimeric transcript). The blue rectangle symbolizes the processed atpI 5′ UTR, with the target of MTHI1 shown in black. Relevant restriction sites Bu (Bsu36I), Bm, (BsmI), S (SnaBI), P (PflMI), and Hc (HincII, where the selection cassette was inserted) are indicated. Arrows above the map indicate the position of the deletions, while the bottom insert shows the mutation introduced in the MTHI1 binding site (underlined) in the dIMf strain, with mutated nucleotides shown in red. A PvuII site introduced as a restriction fragment length polymorphism marker is boxed.
(D) Accumulation of the chimeric petA transcript in the {aAdI} strain transformed with the indicated dIf variants, assessed by RNA gel blots. Three independent transformants are shown for each construct. psaB mRNA is shown as a loading control.
(E) Accumulation of the chimeric cytochrome f (cyt. f) in the same strains, and in the {aAdI}, mthi1-1 {dIf}, and ΔpetA strains as controls, assessed by immunoblots. Immuno-detection of OEE2 is provided as a loading control.