Figure 6.
Disruption of βC1 Binding to GAPCs Reduces CLCuMuV-Induced Autophagy and Enhances Viral Infection.
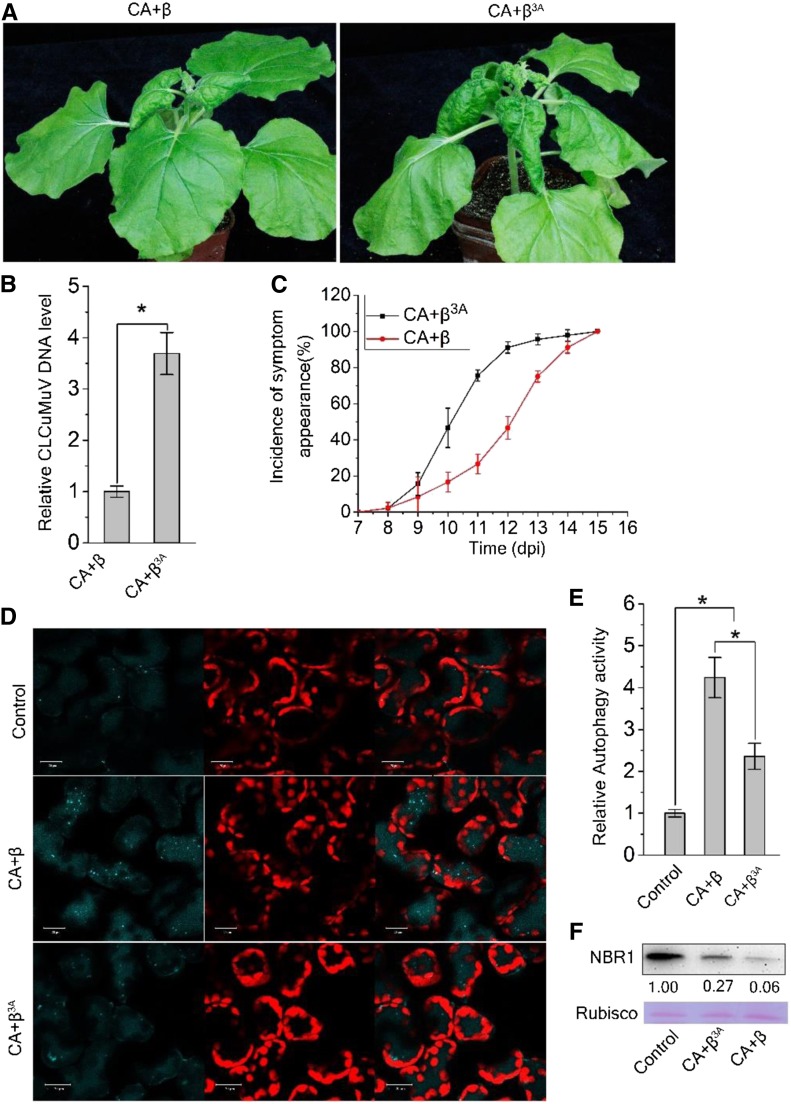
(A) Mutant CLCuMuB (β3A), which encodes a mutant βC13A, caused more severe viral symptoms than wild-type CLCuMuB (β) when co-infected with CLCuMuV (CA). The photographs were taken at 12 dpi. A 3A point mutation in βC1 (βC13A) eliminates its interaction with NbGAPC.
(B) Relative accumulation of CLCuMuV DNA. Quantitative PCR analysis of the V1 gene sequence of CLCuMuV was used to determine viral DNA levels. Values represent means ± se from three independent experiments; t test used for analyses, P ≤ 0.05 (*).
(C) The incidence of symptom appearance at different time points of post infection. Symptom was indicated as the appearance of curled leaves caused by the infection with CA+ β or CA+β3A.
(D) βC13A-carrying virus has reduced induction of autophagy activity. Representative confocal microscopy images of dynamic autophagic activity revealed by the specific autophagy marker CFP-NbATG8f in plants infected with Control, CA+ β, CA+β3A.
(E) Quantification of the CFP-NbATG8f-labeled autophagic puncta/cell from (D). More than 500 mesophyll cells for each treatment were used for the quantification. Relative autophagic activity in noninfected plants was normalized to that of control plants, which was set to 1.0. Values represent means ± se from three independent experiments. P ≤ 0.05 (*).
(F) Protein gel blot assays showed the NbJoka2 protein level in CA+ β or CA+β3A infected plants. NbJoka2 was detected with anti-NBR1 polyclonal antibody.