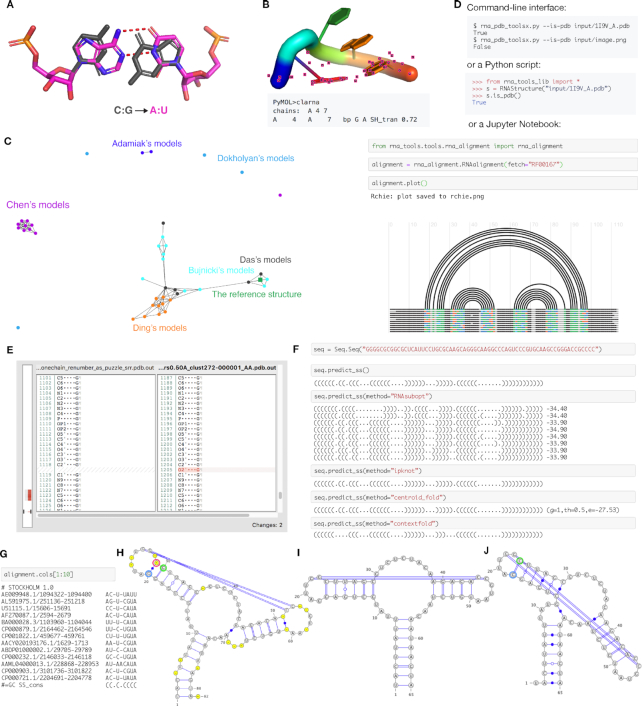
Figure 3.
rna-tools is a set of tools dedicated to RNA structural file manipulation and analysis. (A) Mutate functionality allows for exchanging bases, in this case, C:G pair was replaced with A:U pair from two chains. (B) Contact classification for selected residues can be performed directly in PyMOL. In this case, trans Sugar-Hoogsteen interaction was detected for closing residues of a tetraloop. (C) One of the tools implemented in rna-tools, clanstix. Clanstix can be used for visualizing RNA 3D structures based on pairwise structural similarity (as RMSD) with CLANS. The tool can be used for interactive clustering analysis when various RMSD thresholds can be tested. Here, the clustering of submission for RNA-Puzzle 8 was visualized. Dokholyan submitted four models very different from each other. Models of Chen and of Adamiak were similar respectively and made separate clusters. Models of Ding were similar to each other, and additionally, clustered with models of Das and Bujnicki. When the reference structure was released, it could be added to the visualization. Interestingly, the reference model clustered with two structures of Bujnicki and Das. (D) The functions of the package can be accessed from command-line, from Python scripts, and from Jupyter Notebooks, giving multiple ways to access the functionality. (E) diffpdb checks the consistency between annotations of two structural files. The tool ignores 3D coordinates of atoms and compares only text-content of two files in the PDB to identify the difference in the annotation of atoms, missing atoms (missing the O2′ atom) and missing fragment (shown on the left side with the gray-red bar). (F) Multiple wrappers are implemented allowing for secondary structure prediction performed directly in Jupyter Notebooks, with methods such as RNAsubopt, IPknot, Centroidfold and Contextfold. (G) For RNA alignments it is possible to select only a subset of columns and work on them as a new alignment (in this case on the 1st to the 9th column). Sequences from RNA alignments and their secondary structures can be visualized with VARNA including gaps (H) and without gaps (I). The algorithm checks if residues are ‘paired’ with a gap position (‘−’) (position in red circle) for proper extraction of secondary structure. In this case, after wrong gap removal (J), G (in blue circle) is incorrectly paired with C (in green circle) and all other pairings are shifted by one.