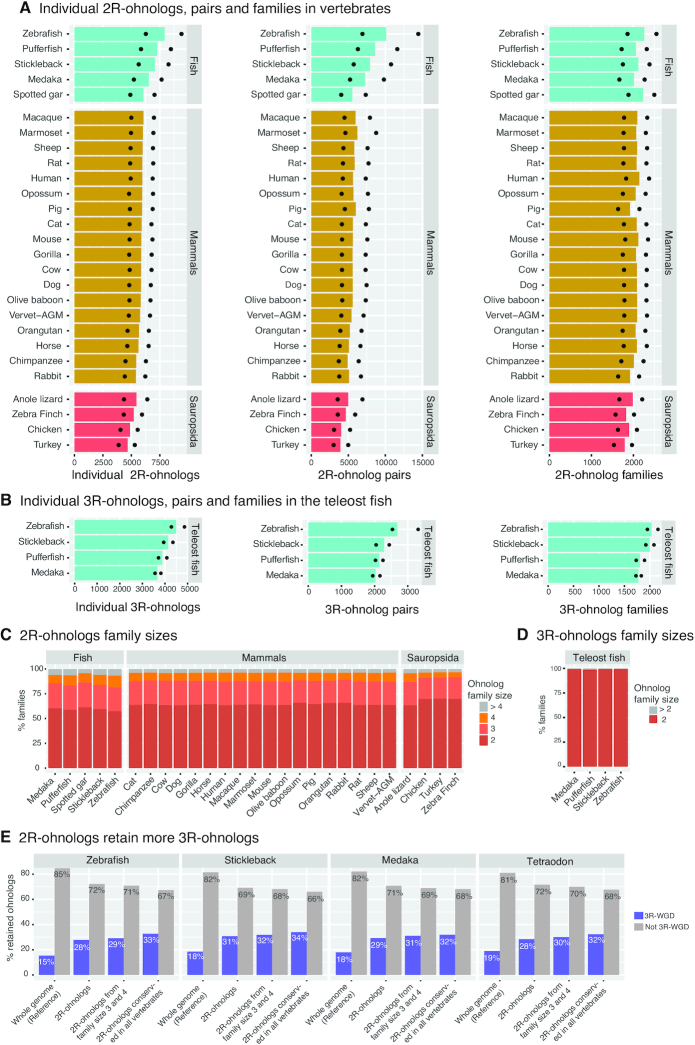
Figure 3.
Description of the ohnolog genes, pairs and families in the database. (A) Number of retained individual 2R-ohnolog genes, pairs and families in all the 27 vertebrates. Bars represent the numbers from the intermediate criterion. Ohnologs from strict and relaxed criteria are indicated by dots. (B) Number of retained individual 3R-ohnolog genes, pairs and families in the four teleost fish species. Bars represent the numbers from the intermediate criterion. Ohnologs from strict and relaxed criteria are indicated by dots. (C) Size of the 2R-ohnolog families from the intermediate criterion in vertebrates. Note that a vast majority of the families are of size 2, 3 or 4. (D) Sizes of the 3R-ohnolog families from the intermediate criterion in the teleost fish hardly exceed size two. (E) The 2R-ohnologs are significantly more likely to retain 3R-ohnologs, compared to genome-average. The retention of 3R-ohnologs is even higher for the 2R-ohnologs that belong to family size 3 or 4, and for 2R-ohnologs conserved in all the 27 vertebrates. All the P-values are <1e-41, Chi-square test. Family counts are from the intermediate criterion.