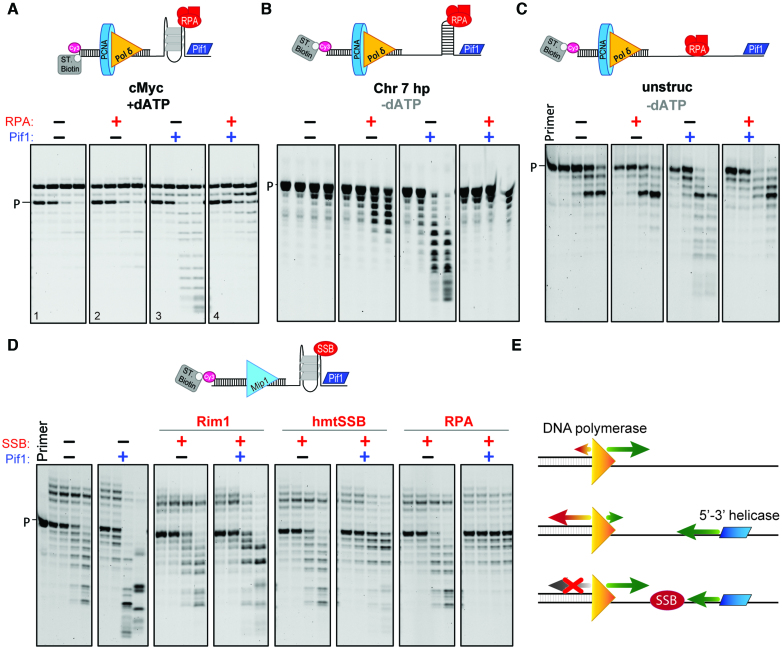
Figure 4.
Head-on polymerase-helicase conflicts result in overt exonuclease activity. (A) Exonuclease digestion of the primer, under conditions of limited DNA synthesis (dATP only), by Pol δ alone (panel 1) or in the presence of either RPA (panel 2) or Pif1 (panel 3) or both (panel 4). The substrate contains cMyc in the template placed 20 nt away from the 3′ end of the primer. (B and C) The same assays but in the absence of DNA synthesis and using DNA substrates that contain either a hairpin sequence or no DNA secondary structures in the template. (D) Exonuclease digestion of the primer by Mip1 alone or in the presence of the indicated combinations of Pif1, ScRim1, human mtSSB, or RPA. (E) Model of helicase-polymerase conflict. In the absence of a 5′-3′ helicase, DNA polymerases favor DNA synthesis over DNA degradation. However, head-conflict between the translocating helicase and the polymerase bound at the 3′-end of the primer results in stimulation of its exonuclease activity. The presence of a single-stranded DNA binding protein (SSB) protects against the helicase-polymerase conflict. Quantification of these gels are shown in Supplementary Figure S14.