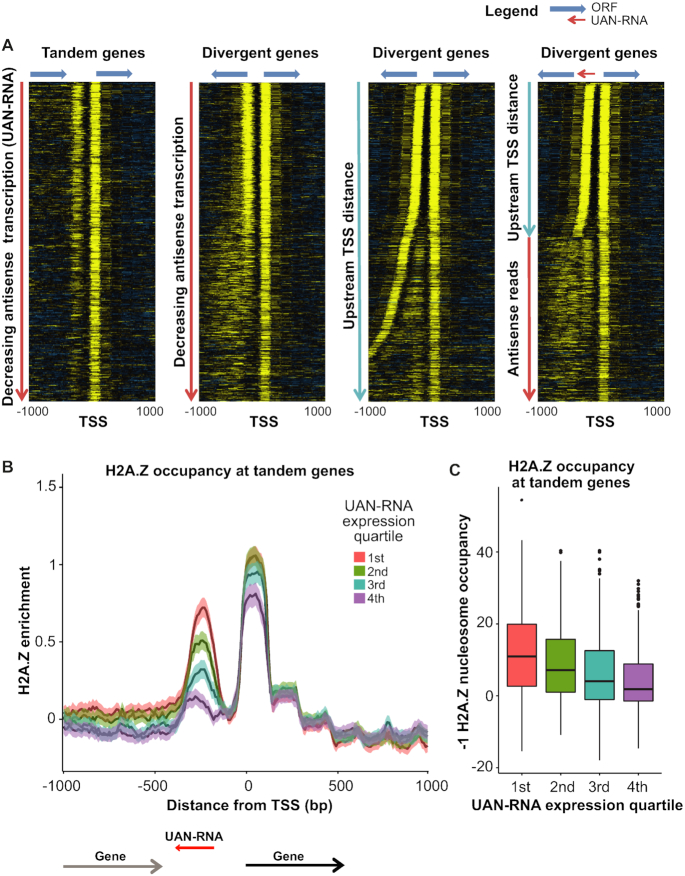
Figure 5.
H2A.Z incorporation at the −1 nucleosome increases with increasing antisense transcription. (A) H2A.Z occupancy data measured by MNase ChIP-seq is plotted for genes segregated based on the orientation of the upstream gene at the TSS. Tandem and divergent genes are first arranged by antisense transcription level in decreasing order (two left panels). The apparent correlation between antisense transcription and H2A.Z incorporation at the −1 nucleosome at divergent genes reflects differences in the intergenic distance between transcript TSS. At divergent genes, it is difficult to distinguish the nucleosome at the 5′ end of the upstream transcript from a nucleosome corresponding to the UAN-RNA transcript. To resolve these, in the second to last heatmap, divergent genes were sorted based on the distance to the upstream TSS. Finally, divergent genes with a TSS-to-TSS distance >350 bp were re-sorted based on the level of UAN-RNA transcription (right-most panel). (B and C) Tandem transcripts were grouped into quartiles according to their UAN-RNA transcription levels. (B) Average profiles are plotted for the quartiles. Overall, H2A.Z occupancy at the −1 nucleosome increases in concert with increasing UAN-RNA transcription level. (C) Boxplots of H2A.Z occupancy at the −1 nucleosome. Welch's t-tests and ANOVA were performed to compare the averages between the four groups, and all comparisons yielded P-values indicating that the differences were significant (P < 2.2 × 10−16).