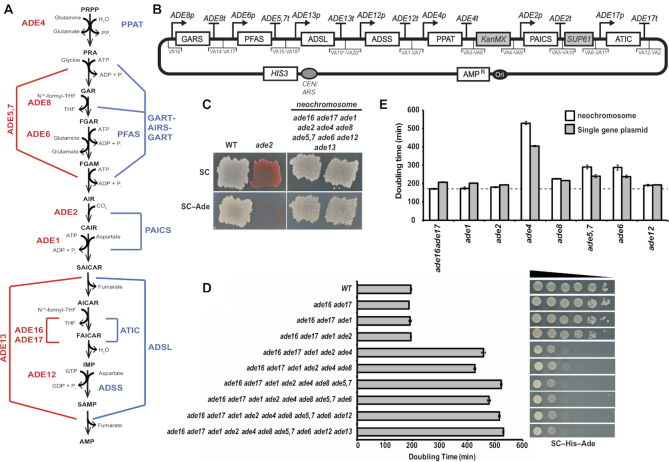
Figure 1.
Yeast and human adenine de novo pathway and humanized strain construction. (A) Schematic representation of the adenine de-novo biosynthesis pathway. Red, yeast genes; blue human. (B) Complete design of the human Purine de-novo Neochromosome for expression in yeast cells. Using yeast golden-gate assembly (18,22) we cloned each human gene (synthesized codon optimized for expression in S. cerevisiae) with its yeast ortholog's promoter and terminator and appropriate adaptors for VEGAS assembly (VA1-VA20) (22) of the neochromosome (asterisk indicate previously unpublished VEGAS adaptors, for details see methods section). (C) Comparing growth on media with and without adenine shows complementation in the humanized strain deleted for all yeast genes and expressing all human genes from the neochromosome. Comparison is to a ade2Δ strain that cannot grow on media without adenine and a WT (wild type; BY4741) that can grow on both. (D) Graphic representation of doubling time in medium without adenine of strains with increasing number of native adenine de novo genes deleted carrying the humanized neochromosome. Grow assay indicates a dramatic increase in doubling time following ADE4 deletion. Spot assay on the left shows a similar trend to the growth assay, showing a dramatic growth defect following deletion of ADE4. (E) Graphic representation of doubling time to verify incomplete complementation of ade4 by its human ortholog PPAT by testing single gene deletions and comparing neochromosome complementation and single gene plasmids in medium without adenine. Dotted line represents doubling time of a wild-type strain grown in the same medium.