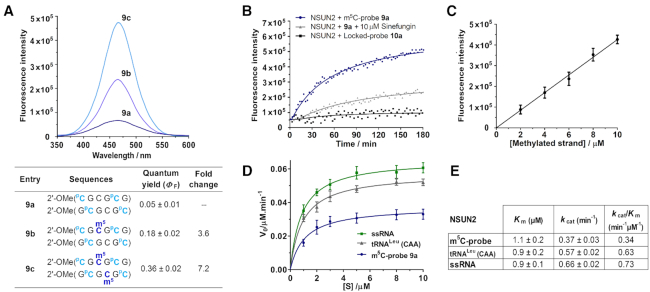
Figure 4.
Verification of the m5C-probe design. (A) The fluorescence emission spectra of probe 9a (λex 360 nm; λem 465 nm) were recorded at 5 μM strand concentration in 10 mM sodium phosphate buffer (pH 7.4) containing 150 mM NaCl and 20 mM MgCl2, 37°C. The probe is highly responsive to its m5C methylation level; introduction of one and two m5C modifications led to a considerable 3.6-fold (9b) and 7.2-fold (9c) increase in fluorescence intensity, respectively. (B) Time-course fluorescence analysis of 9a (5 μM). Significant fluorescence could be detected after a short 30-minute incubation with NSUN2 (0.5 μM; blue line); maximum emission was reached after ∼3 h, giving ∼8 fold increase in fluorescence intensity. Probe fluorescence reduces significantly in the presence of generic MTase inhibitor sinefungin (10 μM; grey line), thus the probe is specifically activated by NSUN2 MTase activity. Incubation of NSUN2 with locked-probe 10a (5 μM; black line) gave negligible fluorescence response, confirming that probe activation is dependent on a change in sugar puckering of PC. (C) The fluorescence intensity increases linearly with the concentration of methylated probe, this enables a direct read-out of NSUN2 MTase activity. Data are expressed as mean ± SD of three replicates. (D, E) Steady-state kinetics analyses of the methylation of m5C-probe, human tRNALeu(CAA) (5′-CCAGACUCAAGUUCUGG-3′) and CpG-rich ssRNA (5′-CGCGCGCGCGCG-3′) by NSUN2. Data are expressed as mean ± SD of three replicates.